Research ArticleGeneticsMuscle biology
Open Access |  10.1172/jci.insight.189286
10.1172/jci.insight.189286
Lack of myotubularin phosphatase activity is the main cause of X-linked myotubular myopathy
Foteini Moschovaki-Filippidou,1 Christine Kretz,1 David Reiss,1 Gaëtan Chicanne,2 Bernard Payrastre,2 and Jocelyn Laporte1
1IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Inserm UMR-S 1258, CNRS UMR7104, Université de Strasbourg, F67404 Illkirch, France.
2I2MC (Institut des Maladies Métaboliques et Cardiovasculaires), Inserm UMR 1297, University of Toulouse, Toulouse, France.
Address correspondence to: Jocelyn Laporte, IGBMC, 1 Rue Laurent Fries, 67404 Illkirch, France. Phone: 0033.388653412; Email: jocelyn@igbmc.fr.
Find articles by Moschovaki-Filippidou, F. in: PubMed | Google Scholar
1IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Inserm UMR-S 1258, CNRS UMR7104, Université de Strasbourg, F67404 Illkirch, France.
2I2MC (Institut des Maladies Métaboliques et Cardiovasculaires), Inserm UMR 1297, University of Toulouse, Toulouse, France.
Address correspondence to: Jocelyn Laporte, IGBMC, 1 Rue Laurent Fries, 67404 Illkirch, France. Phone: 0033.388653412; Email: jocelyn@igbmc.fr.
Find articles by
Kretz, C.
in:
PubMed
|
Google Scholar
|

1IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Inserm UMR-S 1258, CNRS UMR7104, Université de Strasbourg, F67404 Illkirch, France.
2I2MC (Institut des Maladies Métaboliques et Cardiovasculaires), Inserm UMR 1297, University of Toulouse, Toulouse, France.
Address correspondence to: Jocelyn Laporte, IGBMC, 1 Rue Laurent Fries, 67404 Illkirch, France. Phone: 0033.388653412; Email: jocelyn@igbmc.fr.
Find articles by Reiss, D. in: PubMed | Google Scholar
1IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Inserm UMR-S 1258, CNRS UMR7104, Université de Strasbourg, F67404 Illkirch, France.
2I2MC (Institut des Maladies Métaboliques et Cardiovasculaires), Inserm UMR 1297, University of Toulouse, Toulouse, France.
Address correspondence to: Jocelyn Laporte, IGBMC, 1 Rue Laurent Fries, 67404 Illkirch, France. Phone: 0033.388653412; Email: jocelyn@igbmc.fr.
Find articles by Chicanne, G. in: PubMed | Google Scholar
1IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Inserm UMR-S 1258, CNRS UMR7104, Université de Strasbourg, F67404 Illkirch, France.
2I2MC (Institut des Maladies Métaboliques et Cardiovasculaires), Inserm UMR 1297, University of Toulouse, Toulouse, France.
Address correspondence to: Jocelyn Laporte, IGBMC, 1 Rue Laurent Fries, 67404 Illkirch, France. Phone: 0033.388653412; Email: jocelyn@igbmc.fr.
Find articles by
Payrastre, B.
in:
PubMed
|
Google Scholar
|

1IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire), Inserm UMR-S 1258, CNRS UMR7104, Université de Strasbourg, F67404 Illkirch, France.
2I2MC (Institut des Maladies Métaboliques et Cardiovasculaires), Inserm UMR 1297, University of Toulouse, Toulouse, France.
Address correspondence to: Jocelyn Laporte, IGBMC, 1 Rue Laurent Fries, 67404 Illkirch, France. Phone: 0033.388653412; Email: jocelyn@igbmc.fr.
Find articles by
Laporte, J.
in:
PubMed
|
Google Scholar
|

Published October 14, 2025 - More info
JCI Insight. 2025;10(22):e189286. https://doi.org/10.1172/jci.insight.189286.
© 2025 Moschovaki-Filippidou et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: November 15, 2024; Accepted: October 2, 2025
-
Abstract
The MTM1 gene encodes myotubularin (MTM1), a phosphatidylinositol 3-phosphate [PI(3)P] lipid phosphatase. Loss-of-function mutations in MTM1 cause X-linked myotubular myopathy (XLMTM), a severe congenital myopathy with no available cure and a poorly understood pathomechanism. The importance of MTM1 enzymatic activity and its PI(3)P substrate in physiology under normal conditions and in XLMTM is unclear. We generated the Mtm1-KI C375S mice in which the endogenous MTM1 was converted to a phosphatase-dead protein. Mutant mice survived a median of 12 weeks and demonstrated progressively impaired motor skills. Observed muscle hypotrophy and reduced force production compared with their WT littermates (~3.9-fold reduction in absolute maximal force) were responsible for these severe phenotypes. A significantly higher level of PI(3)P was found in the muscle of Mtm1-KI C375S mice. Muscle histology and molecular characterization revealed XLMTM hallmarks, with (a) alteration of the mTOR and autophagy pathways correlating with muscle hypotrophy and (b) abnormal myofiber intracellular organization correlating with impaired muscle force. Overall, this study reveals the importance of MTM1 phosphatase activity and related PI(3)P substrate for postnatal muscle maintenance, and it highlights the significance of MTM1 phosphatase activity in the development of X-linked myotubular myopathy.
-
Introduction
X-linked myotubular myopathy (XLMTM), also called X-linked centronuclear myopathy, is a severe congenital muscle disorder that predominantly affects males and occurs in approximately 1 in 50,000 births worldwide (1, 2). Clinically, XLMTM is characterized by significant muscle weakness, muscle hypotrophy, and respiratory distress and most often results in death within the first months or years of life (3–5). Histological hallmarks include smaller muscle fibers with centralized nuclei (6). There is currently no cure for XLMTM, and its pathomechanism remains poorly understood.
Mutations in the MTM1 gene are responsible for XLMTM. These mutations occur throughout the MTM1 gene and are recognized as loss-of-function mutations (7–11). Indeed, the vast majority correlates with a strong decrease or absence of detection of the MTM1 protein (12).
The MTM1 gene codes for the protein myotubularin (MTM1) that defined a large family of proteins conserved through evolution, with several paralogs mutated in peripheral neuropathies (13–16). MTM1 contains protein-protein and protein-lipid binding domains as the PH-GRAM, Rac-induced recruitment domain (RID), SET-protein interaction domain (SID), and coiled-coil and PDZ-binding domains, and it also contains a phosphoinositides phosphatase catalytic domain (7, 14, 17–22). Mutation of the catalytic cysteine to serine (C375S) results in a complete loss of its enzymatic activity (23, 24). Under normal physiological conditions, MTM1 dephosphorylates phosphatidylinositol 3-phosphate [PI(3)P] to phosphatidylinositol (PI) and phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2] to phosphatidylinositol 5-phosphate [PI(5)P], contributing to the maintenance of phosphoinositides balance within cells (23–25). Through its different protein domains, MTM1 plays a crucial role in membrane trafficking and remodeling and in excitation-contraction coupling (26–29).
Several animal models lacking the MTM1 protein were previously generated and characterized. In mice, 3 different mouse lines were generated through either a KO approach (Mtm1–/y) (30), a gene trap (Mtm1gt/y) (31), or a knock-in of a patient mutation (Mtm1 p.R69C) (32), all leading to the loss of MTM1. As the first mouse model created at IGBMC (Illkirch, France), the Mtm1–/y mouse is a faithful model for XLMTM, accurately reproducing the muscle weakness and histology patterns observed in patients. These mice typically survive approximately until 7 weeks of age. Already at 3 weeks, mice develop a progressive and generalized myopathy with lower body mass (30, 33).
Multi-omics analysis at different time points and in different genetic backgrounds indicates defects in muscle contraction, sarcomere organization, and cell adhesion as potential disease causes and highlights the significance of the abnormalities in neuromuscular junction and the dysregulations in basement membrane pathways (33, 34). Additionally, a recent study has elegantly highlighted the crucial role of MTM1 in maintaining mitochondrial function by coordinating the ER and mitochondria dynamics through PI(3)P regulation on endosomes (35). Finally, overactivation of mTOR and the effect on the LC3 and p62 protein levels in the muscles of Mtm1–/y mice has led to the hypothesis of the mutation suppressing autophagy (31, 36). Dynamin 2 (DNM2) and amphiphysin 2 (BIN1) are also proteins involved in membrane dynamics, are mutated in autosomal forms of centronuclear myopathy and have been implicated in the same molecular pathway together with MTM1 (37–40). In the muscles of Mtm1–/y mice, both DNM2 and BIN1 protein levels are elevated while downregulation of DNM2 and further upregulation of BIN1 rescues Mtm1–/y mice from the myopathic phenotypes (41, 42).
Overall, the importance of the phosphatase activity of MTM1 in muscle physiology and in the XLMTM pathology is still unclear. In particular, apparent discrepancies have been reported. While the Mtm1–/y mouse can be efficiently rescued by gene replacement with exogenous WT MTM1, most of the phenotypes are also rescued with the MTM1-C375S phosphatase-dead or a second phosphatase inactive mutant MTM1-S376N (43). Conversely, Mtm1–/y mice can also be rescued by decreasing the production of PI(3)P through genetic crosses with mice either lacking the class II PI 3-kinase PIK3C2B (44) or specifically lacking the enzymatic activity of PIK3C2B (45), or through treatment with wortmannin, a broad inhibitor of PI 3-kinases (46)
Here, we aimed to resolve this controversy and investigate the importance of MTM1 phosphatase activity in XLMTM. For this purpose, we developed a knock-in mouse model that ubiquitously expresses phosphatase-dead MTM1 (C375S) in place of WT MTM1. We performed a full characterization of this mouse model through behavioral, motor, physiological, histological, ultrastructural, and molecular assessments and deciphered the molecular pathways specifically linked to the regulation of phosphoinositides level by MTM1.
-
Results
Generation and validation of a mouse model lacking MTM1 phosphatase activity. We aimed to develop a novel mouse model expressing MTM1 that lacks phosphatase activity. Since the Mtm1 gene is located on the X chromosome, all our experiments were conducted on male mice. To create the model, we introduced the C375S mutation in the phosphatase domain by replacing the catalytic cysteine with a serine (Figure 1A). Additionally, a silent point mutation was included to facilitate genotyping of the mice by creating a XhoI restriction site (Figure 1, B and C). The presence of these mutations was confirmed by Sanger sequencing (Figure 1B). To validate this model, we firstly demonstrated that MTM1 protein and mRNA levels in muscles were not affected by the mutation and were similar between WT and Mtm1–knock-in C375S mice at 8 weeks of age (Figure 1, D and E). Additionally, MTM1 protein levels were evaluated at a later age of 12 weeks, confirming the presence and stability of the C375S mutant protein over time (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.189286DS1). The subcellular localization of the mutant protein appeared broadly similar to that of the WT, with no major detectable changes based on our qualitative assessment (Supplemental Figure 1B). Nevertheless, the PI(3)P levels were significantly higher in the muscles of Mtm1-KI C375S mice compared with WT mice (almost 2.5-fold increase in the tibialis anterior (TA) and 2-fold increase in the gastrocnemius), confirming the catalytic inactivity of MTM1 in mutant mice, although contributions from other cell types within the muscle cannot be fully excluded (Figure 1, F and G). No differences were observed in the PI3KC2b mRNA levels in the studied muscles, while there was a significant downregulation in the Mtmr1, Mtmr2 and Mtmr5 but not Mtmr13 mRNA levels in the TA muscles of Mtm1-KI C375S mice (Supplemental Figure 8). These data confirm the Mtm1-KI C375S mouse expresses a normal level of phosphatase-dead MTM1.
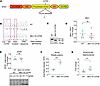 Figure 1
Figure 1Generation and validation of the Mtm1-KI C375S mouse. (A) Representation of the MTM1 protein with its domains, the position of the C375S mutation, and the amino acid sequence around this position. Highlighted in red the catalytic cysteine, replaced by a serine in the mutant. (B) Chromatograms of WT and Mtm1-KI C375S mice. The red boxed indicate the mutation sites and the red arrows the silent mutation introduced for genotyping purposes. (C) Example of genotyping after PCR products were digested with the XhoI enzyme. WT band at 631 bp and Mtm1-KI C375S band at 461 and 170 bp. (D) Mtm1 mRNA levels in the tibialis anterior (TA) muscle of 8-week-old WT and Mtm1-KI C375S mice (n = 6). (E) MTM1 protein levels in the TA muscle of 8-week-old WT and Mtm1-KI C375S mice. WT, n = 8; Mtm1-KI C375S, n = 6. (F) PI(3)P levels in the TA muscle of Mtm1-KI C375S mice and their WT littermates at 8 weeks of age as measured by mass spectrometry. WT, n = 4; Mtm1-KI C375S, n = 3. (G) PI(3)P levels measured by a commercial ELISA kit in the gastrocnemius muscle of 8-week-old WT and Mtm1-KI C375S mice. WT, n = 8; Mtm1-KI C375S, n = 7. Mann-Whitney 2-tailed test for MTM1 protein and PI(3)P levels measured by ELISA; unpaired 2-tailed t test for Mtm1 mRNA levels and PI(3)P levels measured by mass spectrometry; **P ≤ 0.01, ***P ≤ 0.001. Data are shown as mean ± SEM.
Mtm1-KI C375S mice present a decreased survival and a progressive myopathy. Having validated the expression of phosphatase-dead MTM1 by Mtm1-KI C375S mice, we investigated the effect of the specific loss of the phosphatase activity while nonenzymatic functions of MTM1 were conserved. Mtm1-KI C375S mice survived a median of 12 weeks while the longest survivor reached 15 weeks of age (Figure 2A). Though the lifespan of mutant mice was significantly reduced compared with that of WT animals, it should be noted that it was longer than what has been reported for Mtm1–/y mice, which — depending on the genetic background — survive a median of approximately 5–7 weeks (30, 33, 38, 41, 47–51). For assessing the disease progression, we monitored the mice from 4 to 8 weeks of age and evaluated their motor abilities and general clinical condition at weekly intervals. By the age of 4 weeks, the mice exhibited a high disease severity score, indicating motor disabilities, reduced body mass, and the presence of spinal curvature, further supporting that the Mtm1-KI C375S mice were affected by the lack of phosphatase activity. Over the next 4 weeks, their scores increased, pointing to worsening health and disease progression (Figure 2B and Supplemental Video). When 4 weeks old, Mtm1-KI C375S mice showed a 1.16-fold decrease in their total body mass compared with their WT littermates. By 8 weeks of age, this difference increased to 1.26-fold, with the Mtm1-KI C375S mice reaching an average total body mass of 18.6 grams (Figure 2C). Notably, Mtm1–/y mice are well documented to plateau at around 15 gr (30, 33, 38, 47, 48). When 8 weeks old, the mice underwent a total body composition analysis by Nuclear Magnetic Resonance that revealed lower fat and lean mass in comparison to their WT littermates (Figure 2E). These data indicate that a generally altered body composition is a major contributing factor to the lower body mass. Throughout the 4-week study period, Mtm1-KI C375S mice consistently underperformed on the hanging test in comparison with their WT littermates, indicating compromised motor function. A noticeable drop in their ability to hang was observed starting at 6 weeks of age, finally leading to a 2.2-fold decrease in the hanging time compared with their performance when 4 weeks old (Figure 2D). These data, supporting their impaired motor abilities especially at the later time points of the study, aligned with the results from the in situ muscle force measurements on the TA muscle, performed at 8-week-old mice. The absolute maximal force production was almost 4 times lower in the Mtm1-KI C375S mice compared with WT animals. The specific maximal force produced by the Mtm1-KI C375S mice was notably lower at an average of 12.8 mN/mg compared with 19.4 mN/mg for the WT mice, though this difference did not reach statistical significance due to high variation within the mutant group. The mutation’s less profound effect on specific force production compared with absolute force production indicates that the force defects in the model primarily result, at least in part, from muscle hypotrophy (Figure 2, F and G). The absolute force frequency curve revealed that mutant muscles have a strong weakness at all stimulation frequencies while normalization on the muscle mass partly reduce the difference (Figure 2, H and I). It should be noted that the specific force produced by the TA muscle in Mtm1-KI C375S mice was substantially higher than previously reported values in Mtm1–/y mice, where, for example, the maximal specific force was recorded at just around 5 mN/mg (38, 41). Finally, to investigate the muscle fatigability, we performed multiple contraction at 40 Hz. The Mtm1-KI C375S do not show a significant drop in the produced force, as — already at the first stimulations — they fail to develop significant force (Figure 2J). Blood and plasma analysis from samples collected at 8 weeks of age showed no differences in the liver enzyme levels and the blood cell counts between Mtm1-KI C375S and WT mice (Supplemental Figures 2 and 3). Taken together, these results support that mice expressing the MTM1 protein orphaned of its phosphatase activity have a shortened lifespan and develop progressive muscle weakness, similarly to mice completely lacking the protein but possibly with a lower severity.
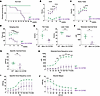 Figure 2
Figure 2Decreased survival and progressive muscle weakness in the Mtm1-KI C375S mouse model. (A) Percentage of survival of Mtm1-KI C375S (n = 9) and WT mice (n = 3). (B–D) Disease severity score, total body mass, and performance on hanging test over 4 weeks of phenotyping, performed weekly, for WT and Mtm1-KI C375S mice. WT, n = 8; Mtm1-KI C375S, n = 7. (E) Fat mass and lean mass of WT and Mtm1-KI C375S mice at 8 weeks of age (n = 5). (F and G) Absolute and specific maximal force measured in situ in the TA muscle stimulated at 150 Hz, in Mtm1-KI C375S mice and their WT littermates at 8 weeks of age. WT, n = 7; Mtm1-KI C375S, n = 6. (H) Absolute TA force measured in situ following different frequency stimulations in 8-week-old Mtm1-KI C375S and WT mice. WT, n = 7; Mtm1-KI C375S, n = 6. (I) Specific submaximal and maximal force of the TA muscle from in situ measurements following stimulations at different frequencies, in 8-week-old Mtm1-KI C375S mice and their WT littermates. WT, n = 7; Mtm1-KI C375S, n = 6. (J) Specific force production by the TA muscle in situ during 80 stimulations at 40Hz, in Mtm 1-KI C375S mice and their WT littermates at 8 weeks of age. WT, n = 7; Mtm1-KI C375S, n = 6. Ordinary 2-way ANOVA with Šídák multiple comparisons for DSS, body mass, hanging test, force-frequency curves and fatigue test; Mann-Whitney 2-tailed test for absolute maximal force; unpaired 2-tailed t test with Welch’s corrections for fat mass, lean mass and specific maximal force; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Data are shown as mean ± SEM.
Muscle hypotrophy and severe structural disorganization in muscles of Mtm1-KI C375S mice. The Mtm1-KI C375S mice present with reduced muscle mass and force, as for patients with XLMTM. We investigated the underlying reason through the characterization of different muscles and of myofibers. No hypotrophy was noted for the gastrocnemius or the quadriceps muscles in Mtm1-KI C375S mice compared with WT controls (Figure 3, A and C). This finding is noteworthy, as the gastrocnemius has previously been reported to exhibit a significantly reduced mass in the Mtm1–/y mouse model when compared with WT animals, even at the early timepoint of 5 weeks (33, 38, 41). The soleus muscles of the Mtm1-KI C375S mice had a significantly smaller mass than that of WT mice, and the normalized weight of the TA muscle in mutant mice showed a 2.1-fold reduction compared with that of their WT littermates (Figure 3, B and D). The absolute muscle mass values are given in Supplemental Figure 4, A–D. As TA was the most affected muscle, we then focused on this muscle for histological analyses.
 Figure 3
Figure 3Muscle and myofibers hypotrophy and histological XLMTM hallmarks in Mtm1-KI C375S mouse model. (A–D) Normalized gastrocnemius, soleus, quadriceps, and TA mass in Mtm1-KI C375S mice and their WT littermates at 8 weeks of age. WT, n = 8; Mtm1-KI C375S, n = 7. (E) Representative pictures of TA muscle sections stained with H&E or SDH from 8-week-old WT and Mtm1-KI C375S mice. Scale bar: 100 μm. (F) MinFeret diameter distribution of TA fibers in Mtm1-KI C375S mice and their WT littermates at 8 weeks of age. WT, n = 5; Mtm1-KI C375S, n = 6. (G) Percentage of large fibers with MinFeret diameter ≥ 40 μm. WT, n = 5; Mtm1-KI C375S, n = 6. (H) Percentage of fibers with mispositioned nuclei in TA muscle sections from 8-week-old WT and Mtm1-KI C375S mice. WT, n = 5; Mtm1-KI C375S, n = 6. (I) Percentage of fibers with abnormal SDH staining in TA muscle sections from 8-week-old WT and Mtm1-KI C375S mice. WT, n = 5; Mtm1-KI C375S, n = 4. Unpaired 2-tailed t test for gastrocnemius, quadriceps, and TA mass; unpaired 2-tailed t test with Welch’s corrections for soleus mass; ordinary 2-way ANOVA with Šídák multiple comparisons for MinFeret distribution curves; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Data are shown as mean ± SEM.
The histological analysis of the TA muscle revealed significant myofiber hypotrophy for the Mtm1-KI C375S mice, with a switch toward smaller fibers and a percentage of large fibers with MinFeret diameter above 40 μm 10 times lower than for the WT mice (Figure 3, E–G, and Supplemental Figure 4E). Furthermore, we found more than 20% of the total fibers analyzed to have mispositioned nuclei, a characteristic XLMTM hallmark (Figure 3, E and H). In the gastrocnemius muscle, histological analysis similarly revealed typical XLMTM hallmarks, although these defects were less pronounced and reached lower statistical significance in all analyzed parameters compared with the TA muscle (Supplemental Figure 5, A–E). These observations appear to correlate with the higher relative increase in PI(3)P levels observed in the TA muscle of mutant mice compared with their WT littermates, which appears more evident than the increase detected in the gastrocnemius (Figure 1, F and G).
Abnormal succinate dehydrogenase (SDH) staining was discovered in more than 5% of the studied fibers, indicating accumulation of oxidative activity, with central or most often subsarcolemmal excessive mislocalization of mitochondria (Figure 3, E and I). Consistent with this result, structural mitochondrial defects and abnormally sized mitochondria were prevalently observed in electron microscopy images of the TA muscles of Mtm1-KI C375S mice (Figure 4). Moreover, the relative mitochondrial/nuclear DNA ratio was 2 times lower in the TA muscles of Mtm1-KI C375S mice compared with their WT littermates (Figure 5A). Nevertheless, ATP production was not significantly affected, while citrate synthase activity was higher in Mtm1-KI C375S mouse TA muscles, indicating an underlying compensatory mechanism at play (Figure 5, B and C). Western blot analysis on the isolated mitochondrial fraction of these muscles showed dysregulated protein levels for complexes of the mitochondrial oxidative phosphorylation system (OXPHOS), additionally indicating damaged mitochondria (Figure 5, D and E). Finally, a 1.3-fold decrease was found for prohibitin protein levels, a marker of the inner mitochondrial membrane (Supplemental Figure 7). No changes were noted in the percentage of type IIa, IIb, or IIx/d within genotypes in the TA muscles, but a significant increase in type I fibers in the TA of mutant animals (Supplemental Figure 6).
 Figure 4
Figure 4Severe structural disorganization in myofibers of Mtm1-KI C375S mice. Representative TA muscle pictures by electron microscopy from 8-week-old WT and Mtm1-KI C375S mice. Mitochondria (M), t-tubules (T-Tub), and triads are indicated in the picture. Scale bar: 1 μm.
 Figure 5
Figure 5Mitochondrial defects in the TA muscles of Mtm1-KI C375S mice. The TA muscles of 8-week-old Mtm1-KI C375S mice and their age-matched WT littermates were analyzed. (A) Mitochondrial DNA to nuclear DNA ratio (n = 4). (B and C) ATP production and citrate synthase (CS) activity as measured by commercial kits (n = 4). (D) Immunoblots demonstrating successful subcellular fractionation based on the relative expression of OXPHOS complexes, GAPDH, and Lamin A. n, nuclear fraction; c, cytoplasmic fraction; m, mitochondrial fraction. Mix of 3 mice per sample for GAPDH and Lamin A. (E) OXPHOS complexes levels in the isolated mitochondrial subcellular fraction (n = 3). Mann-Whitney 2-tailed test for mitochondrial/nuclear DNA ratio, ATP production and OXPHOS CV complex protein levels; unpaired 2-tailed t test for all other comparisons; *P ≤ 0.05, **P ≤ 0.01. Data are shown as mean ± SEM.
Further analysis of the muscle ultrastructure by electron microscopy revealed severely disorganized and smaller sarcomeres with misaligned Z lines and notably lacking normal triad structures (Figure 4). Overall, these findings support that the potentially novel Mtm1-KI C375S mouse model faithfully exhibits all the XLMTM hallmark features in histology, muscle ultrastructure, and mitochondrial defects.
The phosphatase activity of MTM1 is required for normal muscle DNM2 and BIN1 protein levels. Considering the significance of DNM2 protein levels in the XLMTM pathology and the role of BIN1 as a negative regulator of DNM2 (39), we next investigated this pathway in the muscles of Mtm1-KI C375S mice. Similar to observations in the Mtm1–/y mice and in patients with XLMTM (38), we noted significantly higher DNM2 protein levels in the TA muscles of Mtm1-KI C375S mice compared with WT mice (2.2-fold), while the Dnm2 mRNA levels remained unchanged (Figure 6, A–C). BIN1 protein and mRNA levels were 1.9-fold and 1.7-fold higher, respectively, in the muscles of the mutant mice (Figure 6, A, D and E). This is also consistent with findings in Mtm1–/y mice (39). Similar results were observed in the gastrocnemius muscles of Mtm1-KI C375S mice (Supplemental Figure 5, F and G). Taken together, it supports the idea that DNM2 plays a causative role in the disease progression, while changes in BIN1 levels are consequential and act as a compensatory mechanism through transcription adaptation. In conclusion, these data show that DNM2 protein level is regulated by the enzymatic activity of MTM1.
 Figure 6
Figure 6DNM2 and BIN1 protein levels are affected in the muscles of Mtm1-KI C375S mice. (A) Representative blots for DNM2 and BIN1 protein levels in the TA muscles of WT mice and their Mtm1-KI C375S littermates at 8 weeks of age. (B and C) DNM2 protein levels and Dnm2 mRNA levels in the TA muscles of 8-week-old WT and Mtm1-KI C375S mice. For protein WT, n = 8; Mtm1-KI C375S, n = 6. For mRNA, n = 6. (D and E) BIN1 protein levels and Bin1 mRNA levels in the TA muscles of 8-week-old WT and Mtm1-KI C375S mice. For protein WT, n = 8; Mtm1-KI C375S, n = 6. For mRNA, n = 6. Mann-Whitney 2-tailed test for DNM2 protein levels; unpaired 2-tailed t test for all other comparisons; ***P ≤ 0.001. Data are shown as mean ± SEM.
Overactivation of mTOR in muscles of Mtm1-KI C375S mice. MTM1 homologs and PI(3)P were previously implicated in the regulation of the mTOR pathway (52). Moreover, previous studies have reported mTOR overactivation and inhibition of autophagy in the skeletal muscles of Mtm1–/y mice (31, 36). Our study on the Mtm1-KI C375S mouse model shows that elevated PI(3)P levels correlate with the overactivation of mTOR in muscle. Specifically, we observed elevated phosphorylation of S6 ribosomal protein and of p70S6K, both downstream targets of mTOR, in the TA muscles of Mtm1-KI C375S mice (Figure 7, A and B). Interestingly, no changes were found in the phosphorylation of 4EBP1, another downstream target of mTOR involved indirectly in protein synthesis by inhibition of elF4E protein (Figure 7C). LC3b II protein levels were significantly higher (almost 7-fold increase) in the TA muscle of mutant mice compared with WT animals (Figure 7, D and G). Finally, p62 protein levels, a commonly marker for autophagic flux, were 3-fold higher in the muscles of Mtm1-KI C375S mice, while mRNA levels were not affected (Figure 7, E–G). p62 protein is known to accumulate when autophagy is inhibited, leading us to hypothesize that the loss of the MTM1 phosphatase activity results in the decrease of p62 degradation and autophagy. Consistent with this, we also observed increased levels of LC3b II, a marker of autophagosome accumulation, further supporting autophagy disruption. Taken together, these results support that high PI(3)P levels following MTM1 inactivation activate mTOR and inhibit autophagy in muscle.
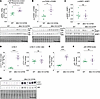 Figure 7
Figure 7mTOR overactivation and autophagy alteration in the muscles of Mtm1-KI C375S mice. (A–C) Protein levels of phosphorylated and total S6 Ribosomal protein phosphorylated and total 70S6K and phosphorylated and total 4EBP1 in the TA muscles of 8-week-old WT and Mtm1-KI C375S mice. WT, n = 8; Mtm1-KI C375S, n = 6. (D) LC3b II protein levels and the LC3b II/LC3b I ratio in the TA muscle of WT and Mtm1-KI C375S mice at 8 weeks of age. WT, n = 8; Mtm1-KI C375S, n = 6. (E and F) p62 protein levels (WT, n = 8; Mtm1-KI C375S, n = 6) and mRNA levels (WT, n = 6; Mtm1-KI C375S, n = 5) in the TA muscles of Mtm1-KI C375S mice and their WT littermates. (G) Western blot images for LC3b I, LC3b II, p62, and total protein. Mann-Whitney 2-tailed test for S6 Ribosomal protein phosphorylation and 4EBP1 phosphorylation; unpaired 2-tailed t test for all other comparisons; *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. Data are shown as mean ± SEM.
-
Discussion
In this study, we generated and characterized the Mtm1-KI C375S mouse, in which the endogenous MTM1 is converted to a phosphatase-dead protein, to distinguish between scaffold and phosphatase-dependent functions and to reveal the role of PI substrate in physiology with a focus on skeletal muscle. We found the MTM1 enzymatic activity and the controlled level of PI(3)P are necessary for survival, motor function, and muscle force, through the regulation of myofiber intracellular organization, organelles positioning, and mTOR activation. While MTM1 is ubiquitously expressed, specific inactivation of its enzymatic activity leads to a progressive myopathy with histological hallmarks similar to XLMTM and to the full MTM1 protein loss in the Mtm1–/y mouse, supporting the loss of the phosphatase activity is the main cause of XLMTM.
MTM1 enzymatic activity and PI(3)P are necessary for myofiber organization and motor functions. Previous myotubularins mutant mice reported to date were created by approaches removing the whole protein but did not allow to discriminate between the scaffold and phosphatase-dependent functions of these enzymes (30–32, 53, 54). The characterization of the Mtm1-KI C375S mouse allowed us, for the first time to our knowledge, to investigate the importance of myotubularins phosphatase activity. Despite its broad tissue expression, MTM1 phosphatase activity and at least its PI(3)P substrate appear mostly essential for skeletal muscle postnatal development and maintenance. In particular, the Mtm1-KI C375S mouse displays muscle hypotrophy and muscle weakness, ultimately leading to a shorter survival. Concerning the role of MTM1 on muscle mass, we propose MTM1 modulates muscle mass through the regulation of PI(3)P and the mTOR and autophagy pathways. Our findings are consistent with studies that have demonstrated the correlation of high PI(3)P levels with mTOR overactivation in muscles, particularly in the context of PI3KC2β inactivation, a kinase producing 3-phosphoinositides (44, 45). Specific inactivation of PI3KC2β kinase activity rescued MTM1 loss in mice (45). Along the same lines, Ebner et al. recently proposed that lysosomal PI(3)P and PI(4)P levels regulate mTOR localization and activity (52). In addition, PI(3)P is a main regulator of autophagy via initiation of the phagophore from endoplasmic reticulum and potentially from sorting endosomes, and via the control of autophagosome-lysosome fusion (55, 56). We cannot exclude other minor phosphoinositides regulated by MTM1, as PI(3,5)P2 or PI(5)P, might also play a role in addition to PI(3)P. Also, DNM2, which is increased following the inactivation of MTM1 phosphatase activity, was implicated in the maturation and recycling of autolysosomes (57). Overall, the control of mTOR and autophagy through the enzymatic activity of MTM1 is necessary to regulate muscle mass.
Concerning the role of MTM1 on muscle force, which is significantly lower in the Mtm1-KI C375S mouse, we hypothesize that PI(3)P regulates myofiber intracellular organization, and especially sarcomere alignment and organelles (mitochondria, triads) position, that are all altered and the main histological defects in the Mtm1-KI C375S mouse. Phosphoinositides in general are implicated in membrane trafficking and cytoskeleton organization. DNM2 also binds membranes, actin, and microtubules (58, 59). In addition, BIN1, which is also higher in the Mtm1-KI C375S muscles, controls T-tubule formation, binds actin and the microtubules end protein CLIP170, and participates to position the nucleus through binding to the nuclear envelope protein nesprin (60, 61). Defects in these intracellular structures lead to inefficient muscle contraction and to the progressive muscle weakness observed in the Mtm1-KI C375S mouse. Overall, the modulation of phosphoinositides by MTM1 enzymatic activity regulates muscle mass and muscle force, while lack of the phosphatase activity triggers muscle hypotrophy and weakness, leading to impaired motor functions and ultimately shorter survival.
The lack of MTM1 phosphatase activity leads to a moderate XLMTM. This study alleviates an apparent controversy in the field of myotubular myopathy. Indeed, MTM1 mutations, even most missense, correlated with an absence of the MTM1 protein, making the importance of the enzymatic activity unclear for disease onset and progression. On one hand, the Mtm1–/y mouse can be efficiently rescued by decreasing the production of PI(3)P through inactivation of the class II PI 3-kinase PIK3C2B, supporting that the enzymatic activity is key (44, 45, 62). On another hand, the Mtm1–/y mouse can be rescued through exogenous expression of phosphatase-dead MTM1 mutants, including C375S mutant, supporting that the scaffolding functions of MTM1 are important (43).
The phenotypes observed in the Mtm1-KI C375S mice clearly demonstrate the manifestation and progression of XLMTM, establishing the Mtm1-KI C375S model as a faithful model for the disease. All XLMTM hallmarks are present in the model, and the molecular pathways affected in the Mtm1–/y were also affected in the Mtm1-KI C375S mice (1, 31, 36). Overall, our model confirms that the disease phenotypes result mainly and directly from the loss of enzymatic activity and from phosphoinositides alteration.
Nevertheless, it is important to note that the present data reveal phenotypes in Mtm1-KI C375S mice that were less severe compared with those previously observed in the previous well-documented Mtm1 mouse models lacking MTM1 under various genetic backgrounds and from different teams (30, 33). Mtm1-KI C375S mice survived longer, reached higher total body mass, and developed greater muscle force with less profound muscle hypotrophy (with no effect on the mass of gastrocnemius). This supports the idea that the scaffolding functions of MTM1 protein influence the severity and progression of the disease but do not affect the establishment of the phenotype. The fact that exogenous expression of MTM1 phosphatase-dead mutants rescued most, albeit not all, XLMTM-like phenotypes in the Mtm1–/y mouse may be due to the effect of scaffolding functions combined to a dominant negative effect on PI(3)P functions (43). Indeed, the exogenous phosphatase-dead MTM1 may still bind its PI(3)P substrate, preventing the deleterious effect of PI(3)P increase in the disease and mimicking the effect of PI(3)P downregulation obtained by PI 3-kinase inhibition. The reported rescue of mitochondrial defects by exogenous expression of MTM1 phosphatase-dead mutants in Mtm1–/y mice in contrast to the presence of these defects in Mtm1-KI C375S mice could be explained by the later timing of exogenous mutant expression (43). These findings indicate that reducing symptom severity in patients with XLMTM could be achieved by mimicking the protein interactions of MTM1, highlighting this approach as a potential direction for future therapy discovery.
-
Methods
Sex as a biological variable. Our study exclusively examined male mice as the Mtm1 gene is on the X chromosome.
Animals. The Mtm1-KI C375S mouse line was established in the Biomedical Sciences Research Centre “Alexander Fleming” in Greece. CRISPR-Cas9 technology was used to insert 3-point mutations in the Mtm1 gene, including codon TGC corresponding to Cys375 to TCG, with the desired mutation resulting in replacing the catalytic cysteine with a serine, and the silent mutation creating an XhoI restriction site for genotyping purposes. Additionally, a silent mutation (T>C) was introduced upstream to the PAM sequence. The crRNA with the sequence GTACTTGTGCACTGCAGTGACGG and the single-stranded oligodeoxynucleotides (ssODN) with sequence 5′-ACCGGTGCCATTCAAGTGGCAGACCAAGTGTCTTCAGGAAAGAGCTCGGTACTTGTGCACTCGAGCGACGGATGGGACAGGACCGCTCAGCTGACATCCTTGGCCATGCTGATGTTGGAC-3′ were used. The line was on a mixed background, 50% C57BL/6J 50% CBA. Mice were housed in the Institut Clinique de la souris animal facility in ventilated cages under controlled conditions of temperature and humidity with 12-hour light/dark cycles, where they had access to food and water ad libitum. Only male littermates were analyzed in this study as the Mtm1 gene is on the X chromosome and as females did not show an obvious phenotype. All experiments were performed on 8-week-old mice, unless otherwise specified.
Genotyping. For genotyping purposes, finger biopsies were used. Genotyping primers were 5′-AGACGGAATGGGAGGTGGT-3′ (forward) and 5′-GGCTTCATTACACTGCTCTTGA-3′ (reverse). The PCR products were digested with the XhoI enzyme for 30 minutes at 37°C. Digestion resulted in a single band with 631 bp size for the WTs and extra bands at 461 and 170 bp for the mutant mice, after electrophoresis of the PCR products. Additionally, the PCR product from one WT and one mutant mouse was subjected to Sanger sequencing for confirmation.
RNA extraction and quantitative PCR. TA and GA muscles were snap frozen in liquid nitrogen and stored at –80°C. Fragments of the muscle samples were used for RNA extraction by lysis in TRl Reagent (Molecular Research Center, #TR 118) and homogenization with a Precellys Evolution Touch homogenizer (Bertin Technologies). For reverse transcription, SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, #18090050) and 1 μg of RNA from the samples were used. For the quantification of mitochondrial to nuclear DNA ratio in TA muscles, DNA was isolated from pieces of the TA muscles using lysis buffer containing 100 mM Tris-HCl, 20 mM NaCl, 0.2% SDS, 5mN EDTA and Proteinase K 100 mg/mL. Samples were incubated at 60°C for 1 hour and final pellets were diluted in TE buffer containing 10 mM Tris-HCl and 1mM EDTA. cDNAs and DNAs were amplified with SYBR Green I Master (Roche, #04887352001) in a LightCycler 480 (Roche). Three technical replicates were utilized for the qPCR. The following primers were used: Mtm1 forward: 5′-TGAGTGAGACTGTCCCTCGG-3′, Mtm1 reverse: 5′-TGGGGCCATTGAAAGGACAT-3′, Nd1 forward: 5′-AAGTTGATCGTAACGGAAGC-3′, Nd1 reverse: 5′-CCCATTCGCGTTATTCTT-3′, Bin1 forward: 5′-CCTGTCTCGCTGCTTGAGAAA-3′, Bin1 reverse: 5′-CTCGAACACCTTCTGGGCTT-3′, Dnm2 forward: 5′-ACCCCACACTTGCAGAAAAC-3′, Dnm2 reverse: 5′-CGCTTCTCAAAGTCCACTCC-3′, p62 forward: 5′-TGTGGAACATGGAGGGAAGA-3′, p62 reverse: 5′-TGTGCCTGTGCTGGAACTT-3′, Mtmr1 forward: 5′-CCTCAACAAGCATGCTTTTCCTCT-3′, Mtmr1 reverse: 5′-CTAGTTGGCACAACAATGATGGCA-3′, Mtmr2 forward: 5′-GACTCACTGTCCAGTGCTTC-3′, Mtmr2 reverse: 5′-TTCTGCTAACTTGTTTGCCTCCC-3′, Mtmr13 forward: 5′-TCTGAAGTACAGTTACCCGTATATC-3′, Mtmr13 reverse: 5′-GATTACATCTAAAAGCTCATGGACATC-3′, Mtmr5 forward: 5′-GAAGTGTTCAGGAATAGCCTTGG-3′, Mtmr5 reverse: 5′-AGCCAATGGGACGGTACATGT-3′, PI3KC2b forward: 5′-CCGACTGGCTACAAAAACACAAC-3′, PI3KC2b reverse: 5′-GCCAGCACAAGAGTAGATGAAGTT-3′, Myh2 forward: 5′-ATCCAAGTTCCGCAAGATCC-3′, Myh2 reverse: 5′-TTCGGTCATTCCACAGCATC-3′, Myh4 forward: 5′-AGACAGAGAGGAGCAGGAGAGTG-3′, Myh4 reverse: 5′-CTGGTGTTCTGGGTGTGGAG-3′, Myh1 forward: 5′-ATGAACAGAAGCGCAACGTG-3′, Myh1 reverse: 5′-AGGCCTTGACCTTTGATTGC-3′, Myh7 forward: 5′-CTACAGGCCTGGGCTTACCT-3′, Myh7 reverse: 5′-TCTCCTTCTCAGACTTCCGC-3′, Rpl27 forward: 5′-AAGCCGTCATCGTGAAGAACA-3′, Rpl27 reverse: 5′ -CTTGATCTTGGATCGCTTGGC-3′, Stau1 forward: 5′-AAGAAGGTGGCCAAGCGTAA-3′, Stau1 reverse: 5′-ATTTGAGTGCTGGCTTGGCA-3′. Rpl27 or Stau1 were used as housekeeping genes for standardization.
Protein extraction and Western blotting. TA and GA muscles were snap frozen in liquid nitrogen and stored at –80°C. Fragments of the muscle samples were lysed in RIPA buffer supplemented with 1 mM PMSF, 1 mM DTT, 5 mM sodium fluoride, 1mM sodium orthovanadate, and complete mini EDTA-free protease inhibitor cocktail (Roche Diagnostics, 11 836 170 001) by incubation for 15 minutes on ice. The tissues were then homogenized using a Precellys Evolution Touch homogenizer (Bertin Technologies) to ensure thorough cell lysis and protein extraction. For some of the experiments, subcellular fractionation of the muscle samples into nuclear, cytosolic, and mitochondrial fractions was performed according to the protocol previously described by Dias et al. (63). Protein concentration in the samples was determined using the DC protein Assay kit (Bio-Rad, 5000113, 5000114, 5000115) according to manufacturer’s instructions. 15 μg of protein were denaturated for 5 minutes at 95°C in 5x Lane Marker Reducing Buffer (Thermo Fisher Scientific) and separated in 10% SDS-PAGE homemade gel or in 4–15% mini-PROTEAN TGX precast protein gel (Bio-Rad, #4561086). PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific, #26619) was also loaded on each gel for reference. Proteins were then transferred to nitrocellulose membranes using a Trans-Blot Turbo Transfer System (Bio-Rad, #1704150). Membranes were stained with Ponceau S to verify protein transfer and to normalize protein loading across samples. Membranes were then blocked in Tris-Buffered Saline with 0.1% Tween-20 (TBS-T) and 5% nonfat dry milk for 1 hour at room temperature and subsequently incubated with primary antibodies overnight at 4°C. The next day the membranes were washed in TBS-T and incubated for 1 hour at room temperature with secondary antibodies. Following the final washes in TBS-T, membranes were visualized with an Ammersham Imager 600 (GE Healthcare Life Sciences) with prior addition of SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, #34577). For the quantification of the band intensity the Fiji software was used. The uncropped pictures of the blots are shown in Supplemental Figure 3. The primary and secondary antibodies used were: anti-MTM1 (rabbit, 1:700, homemade targeting the C-terminal end, #2827) (38), anti-BIN1 (rabbit, 1:1000, R3623; homemade targeting the SH3 domain, #3623), anti-DNM2 (rabbit 1:1000, homemade targeting the proline-rich domain, 2865), anti-prohibitin (rabbit 1:1000, abcam, #ab28172), anti-OXPHOS (mouse 1:1000, Thermo Fisher Scientific, #45-8099), anti-p62 (mouse, 1:1000, Novus Biologicals, #H00008878-M01), anti-phospho-S6 Ribosomal Protein (Ser235/236) (rabbit, 1:1000, Cell Signaling Technology, #2211), anti-S6 Ribosomal Protein (rabbit, 1:1000, Cell Signaling Technology, #2217), anti-phospho-p70S6 (mouse, 1:1000, Cell Signaling Technology, #9206), anti- p70S6 (rabbit, 1:1000, Cell Signaling Technology, #2708), anti-phospho-4EBP1 (rabbit, 1:1000, Cell Signaling Technology, #9459), anti-4EBP1 (rabbit, 1:1000, Cell Signaling Technology, #9644), anti-LC3 (rabbit, 1:1000, Cell Signaling Technology, #9206), anti-GAPDH (mouse, 1:1000, MERCK, #MAB374), anti-Lamin A (rabbit, 1:800, abcam, ab26300), anti-TOMM20 (rabbit, 1:1000, abcam, #ab78547), horseradish peroxidase-coupled goat anti-rabbit (goat, 1:10000, #112-036-04), and horseradish peroxidase-coupled goat anti-mouse (goat, 1:10000, #115-036-068).
PI(3)P mass ELISA. Gastrocnemius muscles were snap frozen in liquid nitrogen and stored at –80°C. To extract the lipids, muscle pieces were homogenized using a Precellys Evolution Touch homogenizer (Bertin Technologies) in 1 mL ice-cold 5% trichloroacetic acid with 1 mM EDTA. The homogenized sample was further diluted in 2 more mL of the same buffer, agitated for 30 seconds and centrifuged for 5 minutes at 900g. The supernatant was discarded and the washing step was repeated another time. The pellet was then resuspended in 3 mL MeOH:CHCl3 and agitated for 10 minutes for the extraction of neutral lipids. After centrifugation for 5 minutes at 900g and a repetition of the neutral lipid extraction step, acidic lipids were extracted by resuspension of the pellet in MeOH: CHCl3: 12 N HCl (80:40:1) and an agitation step of 25 minutes. The samples were then centrifuged and the supernatant was mixed with 0.75 mL CHCl3 and 1.35 mL 1N HCl. A 30-second agitation followed by centrifugation allowed the collection of the lower organic phase from the sample. The organic phase containing the lipids was then dried in a vacuum drier for 1 hour at room temperature. Subsequently, PI(3)P levels were determined in the samples using the PI(3)P mass ELISA kit (Echelon Bioscience Inc, #K-3300) according to manufacturer’s instructions.
Quantification of PI(3)P by mass spectrometry. Phosphoinositides were extracted as described in study by Clark et al. (64) then analyzed as described in study by Li et al. (65). Briefly, 2mg of TA muscle was lysed in lysing matrix S tubes (MP Biomedical) containing 750 μL CHCl3/MeOH/1 M HCl (v/v/v: 10/20/1). Phosphoinositides were extracted with a 2-phase system in which the lower organic phase contained lipids, methylated by (Trimethylsilyl)diazomethane, and separated on Daicel Chiralpak IC-U column for analysis on the LC-QQQ triple quadrupole mass spectrometer (LCMS-8060 Shimadzu) using LabSolutions LCMS software. Offline analysis was performed using LabSolutions Insight LCMS software. Data were quantified as ratio area sample/area internal standard.
Mouse in vivo phenotyping. Starting at 4 weeks of age, mice were monitored daily for severe clinical signs and their body mass was measured. Once a week, the hanging test was performed and the Disease Severity Score (DSS) was assessed. For the hanging test, the mice were suspended upside down on a cage grid for a maximum of 60 seconds. The time of fall was recorder. The test was repeated 3 times per mouse with a 5-minute recovery interval between each trial. The DSS was calculated using a scoring system that evaluated their body mass, their performance on the hanging test, the presence of kyphosis and walking difficulties. Details on the scoring system can been found on Supplemental Table 1. At 8 weeks of age, body composition analysis was performed to determine fat content, lean tissues and free body fluid content. This was achieved using Nuclear Magnetic Resonance (NMR) with the Minispec+ analyzer (LF110-Bruker). Measurements were conducted during light period on conscious, fed mice.
In situ muscle force measurements. Muscle contractile properties were assessed by measuring the contraction of the TA muscle following stimulation of the sciatic nerve. The Complete 1300A mouse Test System (Aurora Scientific) was utilized. Eight-week-old mice were anesthetized by administering of Domitor (2 mg/kg)/Fentanyl (0.28 mg/kg), Diazepam (8 mg/kg), and finally Fentanyl alone (0.28 mg/kg) via i.p. injections. A small incision was made at the lateral side of the leg to expose the sciatic nerve. The distal tendon of the TA muscle was carefully excised and securely tied to the isometric transducer using a nonelastic suture, while the knee and foot were fixed to prevent movement. The sciatic nerve was then stimulated at frequencies ranging from 2 to 150 Hz. Specific force values were calculated by dividing the absolute force by the mass of the TA muscle.
Histology. TA and GA muscles were frozen in liquid nitrogen-cooled isopentane and stored at –80°C. The samples were cut at 8 μm transversal sections and stained with hematoxylin and eosin (H&E) or with succinate dehydrogenase (SDH). The stained samples were then imaged with a Nanozoomer 2HT slide scanner (Hamamatsu) and each myofiber was individually segmented using the Celllpose algorithm (66). To calculate the MinFeret diameter and to manually assess the number of fibers with mispositioned nuclei, Fiji was used. Classification of myofibers regarding SDH staining was done manually with Fiji (67).
Immunofluorescence staining. TA muscles were frozen in liquid nitrogen-cooled isopentane and stored at –80°C. The samples were cut at 8 μm transversal sections. The fiber type immunofluorescence was performed in nonfixed and nonpermeabilized transversal sections. Slides were blocked for 1 hour with 3% BSA in PBS. Antibodies were diluted in 3% BSA in PBS and incubation was performed at 4°C overnight for the primary and at room temperature for 1 hour for the secondary. In the buffer including the secondary antibodies, Wheat Germ Aglutinin Alexa 647 (Invitrogen, # W32466) was also included in a 1:200 dilution. Primary antibodies were anti-myosin type I, BA-D5 (DSHB, AB_2235587), anti-myosin type IIa, sc-71 (DSHB, AB_2147165) and anti-myosin type IIb, BF-F3 (DSHB, AB_2266724), all diluted 1:50. Secondary antibodies were goat anti-mouse IgG2b Cy3 (Jackson ImmunoResearch, #115-165-207), goat anti-mouse IgG1 Alexa 488 (Jackson ImmunoResearch, #115-545-205) and goat anti-mouse IgMμ DyLight 405 (Jackson ImmunoResearch, #115-475-075), all diluted 1:100. Images were acquired using the Axioscan 7 digital slide scanner (ZEISS).
Electron microscopy. Pieces from the TA muscle were fixed in 2.5% paraformaldehyde (PFA, Electron Microscopy Sciences), 2.5% glutaraldehyde (Electron Microscopy Sciences) and 50 mM CaCl2 (Sigma-Aldrich) in cacodylate buffer (0.1M, pH 7.4, Sigma-Aldrich). The samples were then further fixed using 1% osmium tetroxide in cacodylate buffer (0.1M) for 1 hour at 4°C. For dehydration, graded alcohol and propylene oxide were used for 30 minutes each. The samples were then embedded in Epon 812, cut at ultrathin sections (70nm) and contrasted with uranyl acetate and lead citrate. Observation was done under a Philips CM12 electron microscope equipped with a Gatan OneView Camera (Gatan).
ATP production and citrate synthase (CS) activity. ATP levels in TA muscle samples were measured using the ATP Assay Kit (Colorimetric/Fluorometric) (abcam, #ab83355) following the manufacturer’s instructions. Briefly, 10mg of TA muscle were washed in cold PBS and homogenized in 100 μL of the assay buffer provided by the kit, using Dounce homogenizer on ice for 10 passes. The homogenates were then deproteinized using the TCA deproteinization sample preparation kit (abcam, #ab204708) according to manufacturer’s protocol. ATP content was subsequently quantified using the colorimetric assay procedure provided in the kit.
Citrate synthase activity in TA muscles was measured using the Citrate Synthase Assay Kit (abcam, #ab239712) according to manufacturer’s instructions. In short, 10mg of TA muscle were washed in cold PBS and homogenized in 100 μL of the assay buffer provided by the kit using a Precellys Evolution Touch homogenizer (Bertin Technologies). Protein concentration in the samples was determined using the DC protein Assay kit (Bio-Rad, #5000113, 5000114, 5000115) according to manufacturer’s instructions. Samples were transferred in a 96-well plate (dilution factor 10) and reaction mix and background control were added to respective wells. CS activity was measured on a microplate reader in kinetic mode at 412 nm, by following the initial reaction rate of GSH and subsequently CS and CoA levels. Two time points were selected and CS activity was calculated according to the provided formula and normalized to protein levels.
Blood collection and analysis. For the collection of blood and plasma, EDTA-coated Microvette K3E tubes (Sarstedt, #20.1341.100) were used. Samples were kept on ice until analysis with the Element HT5 Hematology Analyzer (SCIL) to determine blood cell counts. For the plasma isolation, samples were centrifuged at 4°C for 15 minutes at 2000g. Plasma was transferred to new tubes and stored at –80°C. Analysis was performed using an AU480 Chemistry Analyzer (Beckman Coulter).
Statistics. All data were verified for normal distribution using the Shapiro-Wilk test. When normality was confirmed, unpaired parametric 2-tailed t test was performed. Welch’s corrections were used when the variances were found unequal. For data not fitting the normal distribution, the Mann-Whitney U test was used. Two-way ANOVA was performed on stimulation frequency to analyze force values during the force frequency protocol, on fiber size to compare percentage of fibers for the H&E staining analysis, on the DSS scores and on the hanging test results, followed by Šídák multiple-comparison test. Statistical significance was considered for P < 0.05. Graphs demonstrate individual points, with additional lines indicating the mean value ± SEM. Statistical analyses were performed in GraphPad Prism 10 software.
Study approval. All animal experiments were performed in accordance with French and European legislations and approved by the institutional ethics committee (project no. APAFIS#35021-2022012717476388).
Data availability. The authors declare that all data supporting the findings of this study are available in the article or its supplemental material. Values for all data points in graphs are reported in the Supporting Data Values file.
-
Author contributions
JL conceived the project. FMF and GC performed molecular experiments. FMF, DR, and CK performed in vivo experiments. JL and BP provided funding and supervised the work. FMF and JL wrote the manuscript.
-
Funding support
Interdisciplinary Thematic Institute IMCBio+, as part of the ITI 2021-2028 program of the University of Strasbourg, CNRS and Inserm.
- IdEx Unistra (ANR-10-IDEX-0002) under the framework of the France 2030 Program.
- SFRI-STRAT’US project (ANR-20-SFRI-0012) under the framework of the France 2030 Program.
- EUR IMCBio (ANR-17-EURE-0023) under the framework of the France 2030 Program.
-
Acknowledgments
The authors would like to thank the scientific platforms at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) and the Institut Clinique de la Souris (ICS) for their help with phenotyping, Nadia Messaddeq for her work on electron microscopy, Mohammed Selloum and Aurelie Auburtin for conducting blood analysis and biochemistry, and Chaouki Bam’hamed for the NMR. Gratitude is also extended to Kostas Bozonelos and the Alexander Fleming Biomedical Sciences Research Center (BSRC) for providing the mutant mouse line [CBA;B6-Mtm1em1(C375S)Flmg/Flmg], and to EMMA/INFRAFRONTIER and BSRC from which the mouse line was distributed. The authors also thank Horia Boursas for her contribution to method development. Finally, the authors would like to thank the Lipidomics core facility of MetaToul and Inserm U1297-I2MC.
Address correspondence to: Jocelyn Laporte, IGBMC, 1 Rue Laurent Fries, 67404 Illkirch, France. Phone: 0033.388653412; Email: jocelyn@igbmc.fr.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2025, Moschovaki-Filippidou et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2025;10(22):e189286.https://doi.org/10.1172/jci.insight.189286.
-
References
- Jungbluth H, et al. Centronuclear (myotubular) myopathy. Orphanet J Rare Dis. 2008;3:26.
- Laporte J, et al. The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet. 2001;17(4):221–228.
- Biancalana V, et al. Characterisation of mutations in 77 patients with X-linked myotubular myopathy, including a family with a very mild phenotype. Hum Genet. 2003;112(2):135–142.
- Barth PG, Dubowitz V. X-linked myotubular myopathy--a long-term follow-up study. Eur J Paediatr Neurol. 1998;2(1):49–56.
- Herman GE, et al. Characterization of mutations in fifty North American patients with X-linked myotubular myopathy. Hum Mutat. 2002;19(2):114–121.
- Romero NB. Centronuclear myopathies: a widening concept. Neuromuscul Disord. 2010;20(4):223–228.
- Laporte J, et al. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13(2):175–182.
- Buj-Bello A, et al. Identification of novel mutations in the MTM1 gene causing severe and mild forms of X-linked myotubular myopathy. Hum Mutat. 1999;14(4):320–325.
- Laporte J, et al. MTM1 mutations in X-linked myotubular myopathy. Hum Mutat. 2000;15(5):393–409.
- Tanner SM, et al. Confirmation of prenatal diagnosis results of X-linked recessive myotubular myopathy by mutational screening, and description of three new mutations in the MTM1 gene. Hum Mutat. 1998;11(1):62–68.
- de Gouyon BM, et al. Characterization of mutations in the myotubularin gene in twenty six patients with X-linked myotubular myopathy. Hum Mol Genet. 1997;6(9):1499–1504.
- Laporte J, et al. Diagnosis of X-linked myotubular myopathy by detection of myotubularin. Ann Neurol. 2001;50(1):42–46.
- Bolino A, et al. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet. 2000;25(1):17–19.
- Raess MA, et al. WANTED - Dead or alive: Myotubularins, a large disease-associated protein family. Adv Biol Regul. 2017;63:49–58.
- Azzedine H, et al. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am J Hum Genet. 2003;72(5):1141–1153.
- Nakhro K, et al. SET binding factor 1 (SBF1) mutation causes Charcot-Marie-Tooth disease type 4B3. Neurology. 2013;81(2):165–173.
- Laporte J, et al. The PtdIns3P phosphatase myotubularin is a cytoplasmic protein that also localizes to Rac1-inducible plasma membrane ruffles. J Cell Sci. 2002;115(pt 15):3105–3117.
- Laporte J, et al. Characterization of the myotubularin dual specificity phosphatase gene family from yeast to human. Hum Mol Genet. 1998;7(11):1703–1712.
- Doerks T, et al. GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem Sci. 2000;25(10):483–485.
- Begley MJ, et al. Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol Cell. 2003;12(6):1391–1402.
- Tsujita K, et al. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J Biol Chem. 2004;279(14):13817–13824.
- Bhattacharyya T, et al. Structural rationale to understand the effect of disease-associated mutations on Myotubularin. Curr Res Struct Biol. 2023;5:100100.
- Blondeau F, et al. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum Mol Genet. 2000;9(15):2223–2229.
- Taylor GS, et al. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci U S A. 2000;97(16):8910–8915.
- Chaussade C, et al. Expression of myotubularin by an adenoviral vector demonstrates its function as a phosphatidylinositol 3-phosphate [PtdIns(3)P] phosphatase in muscle cell lines: involvement of PtdIns(3)P in insulin-stimulated glucose transport. Mol Endocrinol. 2003;17(12):2448–2460.
- Rossi D, et al. Mutations in proteins involved in E-C coupling and SOCE and congenital myopathies. J Gen Physiol. 2022;154(9):e202213115.
- Gómez-Oca R, et al. Common pathogenic mechanisms in centronuclear and myotubular myopathies and latest treatment advances. Int J Mol Sci. 2021;22(21):11377.
- Ketel K, et al. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 2016;529(7586):408–412.
- Kawaguchi K, Fujita N. Shaping transverse-tubules: central mechanisms that play a role in the cytosol zoning for muscle contraction. J Biochem. 2024;175(2):125–131.
- Buj-Bello A, et al. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc Natl Acad Sci U S A. 2002;99(23):15060–15065.
- Fetalvero KM, et al. Defective autophagy and mTORC1 signaling in myotubularin null mice. Mol Cell Biol. 2013;33(1):98–110.
- Pierson CR, et al. Modeling the human MTM1 p.R69C mutation in murine Mtm1 results in exon 4 skipping and a less severe myotubular myopathy phenotype. Hum Mol Genet. 2012;21(4):811–825.
- Sarikaya E, et al. Natural history of a mouse model of X-linked myotubular myopathy. Dis Model Mech. 2022;15(7):dmm049342.
- Djeddi S, et al. Multi-omics comparisons of different forms of centronuclear myopathies and the effects of several therapeutic strategies. Mol Ther. 2021;29(8):2514–2534.
- Jang W, et al. Endosomal lipid signaling reshapes the endoplasmic reticulum to control mitochondrial function. Science. 2022;378(6625):eabq5209.
- Al-Qusairi L, et al. Lack of myotubularin (MTM1) leads to muscle hypotrophy through unbalanced regulation of the autophagy and ubiquitin-proteasome pathways. FASEB J. 2013;27(8):3384–3394.
- Chin YH, et al. Dynamin-2 mutations associated with centronuclear myopathy are hypermorphic and lead to T-tubule fragmentation. Hum Mol Genet. 2015;24(19):5542–5554.
- Cowling BS, et al. Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J Clin Invest. 2014;124(3):1350–1363.
- Cowling BS, et al. Amphiphysin (BIN1) negatively regulates dynamin 2 for normal muscle maturation. J Clin Invest. 2017;127(12):4477–4487.
- Fujise K, et al. Centronuclear myopathy caused by defective membrane remodelling of dynamin 2 and BIN1 variants. Int J Mol Sci. 2022;23(11):6274.
- Tasfaout H, et al. Antisense oligonucleotide-mediated Dnm2 knockdown prevents and reverts myotubular myopathy in mice. Nat Commun. 2017;8:15661.
- Lionello VM, et al. Amphiphysin 2 modulation rescues myotubular myopathy and prevents focal adhesion defects in mice. Sci Transl Med. 2019;11(484):eaav1866.
- Amoasii L, et al. Phosphatase-dead myotubularin ameliorates X-linked centronuclear myopathy phenotypes in mice. PLoS Genet. 2012;8(10):e1002965.
- Sabha N, et al. PIK3C2B inhibition improves function and prolongs survival in myotubular myopathy animal models. J Clin Invest. 2016;126(9):3613–3625.
- Massana-Muñoz X, et al. Inactivating the lipid kinase activity of PI3KC2β is sufficient to rescue myotubular myopathy in mice. JCI Insight. 2023;8(9):e151933.
- Kutchukian C, et al. Phosphatidylinositol 3-kinase inhibition restores Ca2+ release defects and prolongs survival in myotubularin-deficient mice. Proc Natl Acad Sci U S A. 2016;113(50):14432–14437.
- Gómez-Oca R, et al. Differential impact of ubiquitous and muscle dynamin 2 isoforms in muscle physiology and centronuclear myopathy. Nat Commun. 2022;13(1):6849.
- Gayi E, et al. Tamoxifen prolongs survival and alleviates symptoms in mice with fatal X-linked myotubular myopathy. Nat Commun. 2018;9(1):4848.
- Maani N, et al. Tamoxifen therapy in a murine model of myotubular myopathy. Nat Commun. 2018;9(1):4849.
- Al-Qusairi L, et al. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc Natl Acad Sci U S A. 2009;106(44):18763–18768.
- Buono S, et al. Natural history study and statistical modeling of disease progression in a preclinical model of myotubular myopathy. Dis Model Mech. 2022;15(7):dmm049284.
- Ebner M, et al. Nutrient-regulated control of lysosome function by signaling lipid conversion. Cell. 2023;186(24):5328–5346.
- Bolino A, et al. Disruption of Mtmr2 produces CMT4B1-like neuropathy with myelin outfolding and impaired spermatogenesis. J Cell Biol. 2004;167(4):711–721.
- Robinson FL, et al. Loss of the inactive myotubularin-related phosphatase Mtmr13 leads to a Charcot-Marie-Tooth 4B2-like peripheral neuropathy in mice. Proc Natl Acad Sci U S A. 2008;105(12):4916–4921.
- Cebollero E, et al. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012;22(17):1545–1553.
- Puri C, et al. Mammalian autophagosomes form from finger-like phagophores. Dev Cell. 2023;58(23):2746–2760.
- Puri C, et al. A DNM2 centronuclear myopathy mutation reveals a link between recycling endosome scission and autophagy. Dev Cell. 2020;53(2):154–168.
- Gu C, et al. Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 2010;29(21):3593–3606.
- Obar RA, et al. Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature. 1990;347(6290):256–261.
- D’Alessandro M, et al. Amphiphysin 2 orchestrates nucleus positioning and shape by linking the nuclear envelope to the actin and microtubule cytoskeleton. Dev Cell. 2015;35(2):186–198.
- Lee E, et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science. 2002;297(5584):1193–1196.
- Ribeiro I, et al. Phosphoinositide regulation of integrin trafficking required for muscle attachment and maintenance. PLoS Genet. 2011;7(2):e1001295.
- Dias PRF, et al. Subcellular fractionation of frozen skeletal muscle samples. Biochem Cell Biol. 2020;98(2):293–298.
- Clark J, et al. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat Methods. 2011;8(3):267–272.
- Li P, Lämmerhofer M. Isomer selective comprehensive lipidomics analysis of phosphoinositides in biological samples by liquid chromatography with data independent acquisition tandem mass spectrometry. Anal Chem. 2021;93(27):9583–9592.
- Stringer C, et al. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods. 2021;18(1):100–106.
- Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682.
-
Version history
- Version 1 (October 14, 2025): In-Press Preview
- Version 2 (November 24, 2025): Electronic publication











