Research ArticleAIDS/HIVVirology Free access | 10.1172/jci.insight.134196
Cell-associated HIV-1 RNA predicts viral rebound and disease progression after discontinuation of temporary early ART
Alexander O. Pasternak,1 Marlous L. Grijsen,2,3 Ferdinand W. Wit,2,4,5,6 Margreet Bakker,1 Suzanne Jurriaans,7 Jan M. Prins,2 and Ben Berkhout1
1Laboratory of Experimental Virology, Department of Medical Microbiology, and
2Department of Internal Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
3Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom.
4Global Health program, Amsterdam Public Health research institute, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
5Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands.
6HIV Monitoring Foundation, Amsterdam, Netherlands.
7Laboratory of Clinical Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
Find articles by
Pasternak, A.
in:
PubMed
|
Google Scholar
|

1Laboratory of Experimental Virology, Department of Medical Microbiology, and
2Department of Internal Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
3Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom.
4Global Health program, Amsterdam Public Health research institute, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
5Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands.
6HIV Monitoring Foundation, Amsterdam, Netherlands.
7Laboratory of Clinical Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
Find articles by Grijsen, M. in: PubMed | Google Scholar
1Laboratory of Experimental Virology, Department of Medical Microbiology, and
2Department of Internal Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
3Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom.
4Global Health program, Amsterdam Public Health research institute, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
5Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands.
6HIV Monitoring Foundation, Amsterdam, Netherlands.
7Laboratory of Clinical Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
Find articles by Wit, F. in: PubMed | Google Scholar
1Laboratory of Experimental Virology, Department of Medical Microbiology, and
2Department of Internal Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
3Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom.
4Global Health program, Amsterdam Public Health research institute, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
5Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands.
6HIV Monitoring Foundation, Amsterdam, Netherlands.
7Laboratory of Clinical Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
Find articles by Bakker, M. in: PubMed | Google Scholar
1Laboratory of Experimental Virology, Department of Medical Microbiology, and
2Department of Internal Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
3Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom.
4Global Health program, Amsterdam Public Health research institute, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
5Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands.
6HIV Monitoring Foundation, Amsterdam, Netherlands.
7Laboratory of Clinical Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
Find articles by Jurriaans, S. in: PubMed | Google Scholar
1Laboratory of Experimental Virology, Department of Medical Microbiology, and
2Department of Internal Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
3Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom.
4Global Health program, Amsterdam Public Health research institute, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
5Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands.
6HIV Monitoring Foundation, Amsterdam, Netherlands.
7Laboratory of Clinical Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
Find articles by Prins, J. in: PubMed | Google Scholar
1Laboratory of Experimental Virology, Department of Medical Microbiology, and
2Department of Internal Medicine, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
3Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom.
4Global Health program, Amsterdam Public Health research institute, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
5Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands.
6HIV Monitoring Foundation, Amsterdam, Netherlands.
7Laboratory of Clinical Virology, Department of Medical Microbiology, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
Find articles by Berkhout, B. in: PubMed | Google Scholar
Published February 25, 2020 - More info
JCI Insight. 2020;5(6):e134196. https://doi.org/10.1172/jci.insight.134196.
© 2020 American Society for Clinical Investigation
Received: October 14, 2019; Accepted: February 19, 2020
-
Abstract
Plasma viral load (VL) and CD4+ T cell count are widely used as biomarkers of HIV type 1 (HIV-1) replication, pathogenesis, and response to antiretroviral therapy (ART). However, the clinical potential of cell-associated (CA) HIV-1 molecular markers is much less understood. Here, we measured CA HIV-1 RNA and DNA in HIV-infected individuals treated with temporary ART initiated during primary HIV-1 infection. We demonstrate substantial predictive value of CA RNA for (a) the virological and immunological response to early ART, (b) the magnitude and time to viral rebound after discontinuation of early ART, and (c) disease progression in the absence of treatment. Remarkably, when adjusted for CA RNA, plasma VL no longer appeared as an independent predictor of any clinical endpoint in this cohort. The potential of CA RNA as an HIV-1 clinical marker, in particular as a predictive biomarker of virological control after stopping ART, should be explored in the context of HIV-1 curative interventions.
-
Introduction
Combination antiretroviral therapy (ART) can successfully manage but is unable to cure HIV type 1 (HIV-1) infection (1). The main goal of ART is durable suppression of virus replication, measured in clinical settings by the concentration of free virus particles in peripheral blood plasma (plasma viral load, VL), to below the detection limit of the most sensitive clinical assay available. In most individuals adherent to modern ART regimens, this goal of plasma VL undetectability is achieved, but virus is nonetheless never completely eradicated. Therefore, additional virological markers for monitoring the response to therapy can be useful if they are informative, and ideally predictive, of the degree of virological and immunological response to ART, indicating a timely intervention (e.g., ART intensification of a behavioral intervention to improve adherence) if necessary. Indeed, cell-associated (CA) HIV-1 RNA has been demonstrated to predict ART failure and to correlate with adherence to therapy in virologically suppressed individuals (2, 3), indicating that this marker could be of use in clinical care of individuals on otherwise-suppressive ART because it is more sensitive than plasma VL for associations with certain clinical endpoints. Pre-ART plasma VL, CD4+ T cell count, and total HIV-1 DNA have been shown to predict the rates of virological suppression on ART (4–6). However, to the best of our knowledge, the predictive value of CA RNA for the response to ART has not yet been reported. Likewise, CD4+ count and plasma VL are firmly established as independent predictors of disease progression in the absence of treatment (7–9), and studies have reported an independent predictive value of total HIV-1 DNA for disease progression (10–13). However, no study has directly compared plasma VL and CA HIV-1 RNA for the prediction of the rate of CD4+ T cell decline in the setting of untreated HIV-1 infection.
Although viral replication drops to undetectable levels within weeks to months after starting ART, HIV-1 persists in long-lived blood and tissue reservoirs and almost inevitably rebounds if ART is interrupted (14–16). Persistence of viral reservoirs is considered a main obstacle to the development of an HIV-1 cure (17). Recent efforts toward a cure for HIV-1 resulted in a renewed interest in the development of viral markers, which should allow the precise measurement of HIV-1 reservoirs and aid in monitoring the efficacy of novel therapies aimed at eliminating or reducing these reservoirs (18). In particular, because every potential curative intervention will ultimately necessitate an analytical treatment interruption (ATI) to assess its efficacy, it is very important to be able to identify individuals with small viral reservoirs, in whom ATI can be performed with the lowest risk of quick viral rebound, because the latter could result in HIV-1 reservoir replenishment, selection of drug resistance, disease progression, and elevated risk of HIV-1 transmission (19, 20). Only a minority of individuals demonstrate some level of spontaneous HIV-1 control after interruption of ART, which in almost all such cases has been initiated during early infection (21–23). However, the absence of reliable predictive markers of the duration of such ART-free HIV-1 remission (posttreatment control) seriously complicates clinical decision-making about ART interruption and therefore hinders HIV-1 cure research (24). A reliable predictive biomarker could even allow the initial assessment of novel curative interventions without the need for ART interruption. Some candidate predictive biomarkers for the duration of ART-free remission have been proposed (25–27); however, different studies measured different markers, used different thresholds for viral rebound, and performed different types of analysis, which resulted in some contradictory conclusions. As a result, validated biomarkers predictive of virological control after ART interruption are not yet available, which means ATIs are still the best way to assess efficacy of new therapeutic interventions, and criteria to be used in recruitment of participants for HIV-1 curative trials remain unclear.
In the present study, we measured a number of virological markers in peripheral blood mononuclear cells (PBMCs) of participants of the Primo-SHM study, a randomized controlled trial comparing no treatment with 24 or 60 weeks of temporary ART initiated during primary HIV-1 infection (PHI) (28). Temporary ART provided an opportunity to measure HIV-1 markers at several time points in order to assess their predictive value for different clinical endpoints, such as virological and immunological response to ART, viral rebound upon ART interruption, and disease progression in the absence of treatment. Remarkably, we found an independent predictive value of different CA HIV-1 RNA species for each of these endpoints. Surprisingly, when adjusted for CA RNA, plasma VL no longer was an independent predictor of any clinical endpoint in this cohort. These results argue that the potential of CA HIV-1 RNA as a clinical marker for monitoring HIV-1 replication and pathogenesis, as well as the response to ART, deserves further exploration. Perhaps even more importantly, the role of CA RNA as a predictive biomarker of virological control upon stopping ART should be explored in the context of HIV-1 curative interventions.
-
Results
Study participants and variables. All 64 participants of the Primo-SHM trial who completed the trial in the Academic Medical Center of the University of Amsterdam (AMC) were included in this study. Baseline and treatment characteristics of the study participants are shown in Table 1. In brief, the median age was 39.8 years and 95.3% were males. Most participants (82.8%) were MSM, and 84.4% were infected with HIV-1 subtype B. The study design is shown in Figure 1. Participants received 60 weeks’ (n = 29), 24 weeks’ (n = 23), or no (n = 12) early ART.
 Figure 1
Figure 1Schematic of the study. Participants received either 60 weeks’ (n = 29), 24 weeks’ (n = 23), or no (n = 12) early ART. Variables were measured at 3 time points (shown in red): baseline (corresponding to the start of early ART for the 60-week and 24-week arms), early ART interruption (for 60-week and 24-week arms), and virological set point (36 weeks from early ART interruption or baseline). Numbers of participants included in the corresponding analyses: baseline (n = 44), treatment interruption (n = 51), and set point (n = 56).
All virological and immunological variables — plasma VL, CA HIV-1 unspliced (US) (gag) and multiply spliced (MS) (tat/rev) RNA, total HIV-1 DNA, CD4+ count, and CD4/CD8 ratio — were measured at baseline (corresponding to the start of early ART for the 60-week and 24-week arms) and at the virological set point. The latter was defined, in accordance with the original Primo-SHM trial design (28), at 36 weeks from early ART interruption (TI) or, for the no-treatment arm, at 36 weeks from baseline, to allow for stabilization of the plasma VL in untreated and treated patients (29, 30) and to minimize dropout due to rapid disease progression (Figure 1). During early ART, plasma VL, CD4+ count, and CD4/CD8 ratio were measured weekly during the first month, monthly during the second and third months, and every 3 months for the remainder of the period on therapy. After TI, these variables were measured bimonthly for the first month, monthly during the second and third months, and subsequently every 3 months. US and MS RNA and total DNA were measured every 12 weeks throughout the early ART periods. For the present study, measurements of these variables on the latest available visit before TI were included in the analysis as candidate predictors of viral rebound and CD4+ cell loss upon TI. Because MS RNA was rarely detectable during ART, participants were stratified into those with detectable or undetectable MS RNA in any on-ART measurement, and MS RNA detectability on ART was included in this analysis as a categorical variable.
As expected, levels of all virological markers decreased on ART and increased again after TI, while the opposite dynamics were observed for the immunological markers (CD4+ count and CD4/CD8 ratio) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.134196DS1). Detailed analysis of the longitudinal dynamics of virological and immunological markers on early ART, as well as comparisons between baseline and post-TI set point values of the markers per treatment arm will be reported separately. At baseline and at set point, all variables significantly correlated between each other: positive correlations were observed in any pair of virological markers, as well as between CD4+ count and CD4/CD8 ratio, whereas virological markers negatively correlated with immunological markers (Figure 2, A and C). At TI, we only observed (positive) correlations between CD4+ count and CD4/CD8 ratio and between US RNA and total DNA, whereas no significant correlations were detected between virological and immunological markers (Figure 2B). Strong positive correlations were also observed for all virological and immunological markers between the time points (Figure 2D). For each of these 3 time points, the measured variables were assessed as candidate predictors of the corresponding clinical endpoints, as follows: baseline variables were assessed for the prediction of response to ART, TI variables for the prediction of viral rebound and disease progression, and set point variables for the prediction of disease progression. To compare the predictive power of different explanatory variables for the corresponding clinical endpoints, only participants without missing values on any variable (per time point) were included in the corresponding analyses (Figure 1). In addition, a number of demographic and clinical covariates, such as age, sex, HIV-1 transmission route, HIV-1 subtype, Fiebig stage of early infection, early ART duration, NRTI backbone, and ART class, were included in the analyses as appropriate.
 Figure 2
Figure 2Correlation matrices of clinical and virological variables. (A–C) Correlations between different variables measured at (A) baseline (BL), (B) early ART interruption (TI), and (C) virological set point (SP). Participants from all 3 arms were included in the BL and SP correlation analyses; participants from 24- and 60-week treatment arms were included in the TI correlation analysis. Numbers of measurements are shown for every variable and for every time point. (D) Correlations between different time points per variable. Heatmaps indicate the strength of positive (blue) or negative (red) correlations. Spearman’s rho coefficients are depicted below the diagonal and P values are above the diagonal. US RNA, unspliced RNA; MS RNA, multiply spliced RNA.
CA HIV-1 RNA predicts virological and immunological response to ART. First, we assessed the predictive value of baseline variables for the time to achieve virological suppression (plasma VL < 50 copies/mL) on early ART. All participants included in the baseline analysis, except 1, achieved virological suppression on therapy within a median of 115 (IQR, 84–168) days, counting from the baseline to the first undetectable plasma VL measurement. The single participant who did not achieve virological suppression was censored from the analysis at TI. For the univariate Kaplan-Meier analysis, levels of every potential continuous predictor were stratified into “high” and “low” strata based on median values, and times to virological suppression were compared between participants with high and low values of every predictor. In this analysis, among all potential predictors, only plasma VL, US RNA, and total DNA were significantly associated with virological suppression (P = 0.0029, P = 0.0035, and P = 0.0007, respectively) (Figure 3 and Supplemental Table 1). Univariable Cox analysis, in which all continuous variables were included without categorization, revealed that the same variables, as well as CD4/CD8 ratio, were significantly associated with virological suppression (Table 2). However, only CD4/CD8 ratio remained a significant independent predictor in the final multivariable Cox regression model (aHR = 1.14 [1.01–1.30], and P = 0.035). In addition, a trend toward US RNA predicting virological suppression was observed (aHR = 0.72 [0.51–1.02], and P = 0.063) (Table 2).
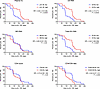 Figure 3
Figure 3Pre-ART predictors of time to virological suppression on early ART. Kaplan-Meier survival analysis for plasma VL, CA HIV US RNA, CA HIV MS RNA, total HIV DNA (VD), CD4+ count, and CD4/CD8 ratio, measured at the start of early ART (n = 44). The endpoint of the analysis was virological suppression less than 50 copies/mL. Levels of every potential predictor were stratified into “high” and “low” strata based on median values. HRs and 95% CIs were calculated using Mantel-Haenszel method. P values were calculated using log-rank tests.
 Table 2
Table 2Baseline variables associated with time to achieve virological suppression (plasma VL < 50 copies/mL) on early ART (n = 44; Cox proportional hazards analysis)
We then asked whether any of the baseline variables would be predictive for virological suppression at an early time point after ART initiation. Levels of every potential predictor were compared between participants who did and did not achieve virological suppression at 12 weeks of ART by Mann-Whitney U or Fisher’s tests. In this analysis, only US RNA was significantly associated with virological suppression (P = 0.032), whereas plasma VL was not predictive (Figure 4 and Supplemental Table 2). The same result was obtained by univariable and multivariable logistic regression analyses (adjusted odds ratio [aOR] = 0.48 [0.25–0.93], P = 0.029) (Table 3).
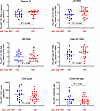 Figure 4
Figure 4Pre-ART predictors of virological suppression at 12 weeks of early ART. Plasma VL, CA HIV US and MS RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio, measured at the start of early ART, were compared between participants with undetectable (n = 19, blue dots) and detectable (n = 24, red dots) plasma VL at 12 weeks of early ART. P values were calculated using Mann-Whitney U tests. Open circles depict undetectable values, censored to 50% of assay detection limits.
 Table 3
Table 3Baseline variables associated with virological suppression (plasma VL < 50 copies/mL) at 12 weeks of early ART (n = 43; logistic regression analysis)
Having shown the predictive value of CA RNA for virological response to early ART, we next investigated whether it would also be predictive for the immunological response to therapy. Normalization of CD4/CD8 ratio to more than 1 is considered an important measure of immunological response to ART, and a low CD4/CD8 ratio is a prognostic marker for both opportunistic infections and non-AIDS morbidity and mortality (31, 32). Because many participants, especially those who were treated for 24 weeks, did not achieve a CD4/CD8 ratio of more than 1 by the time of TI, we did not consider survival analysis appropriate because all these participants would have been censored, which would have biased the results. Instead, we assessed the predictive power of the baseline variables for the normalization of CD4/CD8 ratio in the first year of early ART (only the 60-week treatment arm was included in this analysis). We compared levels of every potential predictor between participants who did and did not achieve a CD4/CD8 ratio of more than 1 at 48 weeks of ART. In addition to the baseline CD4/CD8 ratio itself (P = 0.039), only MS RNA was significantly associated with immunological response (P = 0.0043) (Figure 5 and Supplemental Table 3). In the univariable and multivariable logistic regression analyses, only MS RNA was significantly predictive of immunological response to ART (aHR = 0.14 [0.03–0.73], P = 0.020), whereas baseline CD4/CD8 ratio was not predictive (Table 4).
 Figure 5
Figure 5Pre-ART predictors of CD4/CD8 ratio normalization at 48 weeks of early ART. Plasma VL, CA HIV US and MS RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio, measured at the start of early ART, were compared between participants with CD4/CD8 ratio less than 1 (n = 8, blue dots) and more than 1 (n = 16, red dots) at 48 weeks of early ART. P values were calculated using Mann-Whitney U tests. Open circles depict undetectable values, censored to 50% of assay detection limits.
 Table 4
Table 4Baseline variables associated with CD4/CD8 ratio more than 1 at 48 weeks of early ART (n = 24; logistic regression analysis)
CA HIV-1 RNA predicts the magnitude of viral rebound after TI. All individuals who interrupted early ART (n = 51) achieved viral rebound to more than 50 copies/mL within 252 days and to more than 400 copies/mL within 574 days, counting from the moment of TI to the first plasma VL measurement higher than the corresponding threshold (Supplemental Figure 2). The median magnitude of viral rebound, measured as plasma VL at the virological set point (36 weeks after TI) was 4.43 (IQR, 3.63–4.94) log10 copies/mL. We first assessed the predictive power of variables measured before TI for the magnitude of viral rebound. In both uni- and multivariable analysis, US RNA was the only variable significantly associated with plasma VL at set point (P = 0.003, and P = 0.020, respectively) (Figure 6A). We reasoned, however, that this association might be partly explained by the predictive power of pre-ART virological markers for viral rebound. Although pre-ART CD4+ count, CD4/CD8 ratio, US RNA, and plasma VL were all significantly associated with viral rebound in the univariable model, only US RNA remained as an independent predictor in the multivariable model (P = 0.001). In addition, pre-ART total HIV-1 DNA, which was not associated with viral rebound in the univariable model, became strongly negatively associated in the multivariable model (P = 2.9 × 10–4) (Figure 6B). Because the latter 2 pre-ART variables were strongly positively correlated (rho = 0.65, P = 7.7 × 10–8) (Figure 2A), the apparent lack of predictive power of total DNA for viral rebound in the univariable model can be explained by masking of this effect by the high positive predictive power of US RNA. Controlling for US RNA in the multivariable model revealed the negative predictive power of total DNA.
 Figure 6
Figure 6Predictors of magnitude of viral rebound after interruption of early ART. (A and B) Coefficient plots of variables measured (A) at early ART interruption (n = 51) or (B and C) at the start of early ART (baseline) (n = 43). Effect sizes and 95% CIs for the associations of the variables with plasma VL at the virological set point were calculated by fitting univariable and multivariable generalized linear models. For baseline, 2 models are shown, including either US RNA and total HIV-1 DNA separately (model I, B) or the US RNA/total DNA ratio as a single variable (model II, C). US RNA/total DNA ratio was calculated taking into account that 106 PBMCs contain 1 μg of total RNA (74). (D) The final multivariable model. Units of measurement of total HIV-1 DNA are log10 copies/106 PBMCs and of US are log10 copies/μg total RNA. Significant associations are marked in red.
Given the opposite associations of pre-ART US RNA and total DNA with viral rebound, we attempted to further improve the multivariable model by introducing the US RNA/total DNA ratio into the model as a single variable (Figure 6C). Indeed, the association of the pre-ART US RNA/total DNA ratio with viral rebound was stronger than that of either US RNA or total DNA alone, and its inclusion in the multivariable model resulted in an improved model fit (Akaike Information Criterion, 97.8 vs. 102.8). In this improved model, only the US RNA/total DNA ratio was independently predictive of viral rebound (P = 7.5 × 10–5). Pre-ART and TI variables that were predictive of viral rebound were subsequently selected for the final multivariable model (Figure 6D). In this analysis, the associations of TI US RNA and pre-ART US RNA/total DNA ratio with the magnitude of viral rebound remained significant (B = 0.35 [95% CI = 0.09–0.60], P = 0.007; and B = 0.62 [0.37–0.87], P = 1 × 10–6, respectively).
CA HIV-1 RNA predicts the time to viral rebound and CD4+ T cell decline after TI. Median times to viral rebound of more than 50 and more than 400 copies/mL were 35 (IQR, 28–56) days and 55 (IQR, 28–60) days, respectively (Supplemental Figure 2). Because no pre-ART virological marker was predictive for time to viral rebound after TI (Supplemental Table 4), we assessed the predictive power of variables measured before TI for the time to viral rebound. Among the clinical and virological variables, only high US RNA level was significantly associated with shorter time to viral rebound of both more than 50 and more than 400 copies/mL (HR = 0.41 [0.21–0.78], P = 0.0070; and HR = 0.36 [0.18–0.70], P = 0.0026, respectively) (Figure 7). This was confirmed in a subset of participants with plasma VL less than 50 copies/mL at the measurement time points (n = 44) (Supplemental Figure 3). In addition, younger age was associated with longer time to viral rebound of more than 50 but not of more than 400 copies/mL (P = 0.020), and HIV-1 subtype B was marginally associated with shorter time to viral rebound of both more than 50 and more than 400 copies/mL (P = 0.062, and P = 0.059, respectively) (Supplemental Table 5). Surprisingly, longer time on ART (60 vs. 24 weeks) was not associated with longer time to viral rebound, and a trend in the opposite direction was observed, as the 24-week arm demonstrated longer times to viral rebound of more than 400 copies/mL (P = 0.10). The predictive power of TI US RNA was confirmed in the univariable and multivariable Cox analyses because it was the only variable significantly associated with time to viral rebound of both more than 50 and more than 400 copies/mL (aHR = 1.47 [1.02–2.11], P = 0.040; and aHR = 1.47 [1.00–2.14], P = 0.047) (Table 5).
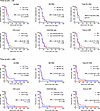 Figure 7
Figure 7On-ART predictors of time to viral rebound after interruption of early ART. Kaplan-Meier survival analysis for CA HIV US RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio, measured at the latest time point available before interruption of early ART, as well as detectability of CA HIV MS RNA at any time point on early ART, and time on early ART (60 or 24 weeks) (n = 51). The endpoint of the analysis was viral rebound to more than 50 or more than 400 copies/mL. Levels of US RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio were stratified into “high” and “low” strata based on median values. HRs and 95% CIs were calculated using Mantel-Haenszel method. P values were calculated using log-rank tests. det, detectable; ud, undetectable.
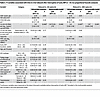 Table 5
Table 5TI variables associated with time to viral rebound after interruption of early ART (n = 51; Cox proportional hazards analysis)
We next sought to establish the predictive role of variables measured before TI for subsequent disease progression. The latter was protocol-defined as the time to experience a drop in the CD4+ count below 350 cells/mm3 or to restart ART. All except 3 participants achieved this composite endpoint by the end of the observation period within a median of 555 (IQR, 102–1350) days from TI. These 3 participants were censored from the analysis at the end of the observation period. Of the remaining 48 participants, 35 reached the CD4+ count of less than 350 cells/mm3, and 13 restarted ART with higher CD4+ counts (by the trial protocol, participants had to start or restart ART in the case of 2 consecutive CD4+ counts less than 350 cells/mm3, symptomatic HIV-1 disease, or physician or participant insistence on starting or restarting ART). In addition to high CD4+ count (HR = 3.12 [1.64–5.93], P = 0.0005), undetectable MS RNA on ART (HR = 0.16 [0.05–0.53], P = 0.0027) and low total HIV-1 DNA at TI (HR = 0.46 [0.25–0.85], P = 0.014) were significantly associated with longer disease progression, while low US RNA was marginally significant (HR = 0.55 [0.30–1.00], P = 0.052) (Figure 8). In addition, HIV-1 subtype B was associated with shorter disease progression (HR = 2.47 [1.29–4.72], P = 0.0062) (Supplemental Table 6). Again, time on ART (60 vs. 24 weeks) was not significantly associated with disease progression. These associations were supported by the univariable Cox regression analysis (Table 6), but only CD4+ count and US RNA remained as independent predictors in the final multivariable Cox model (aHR = 0.65 [0.55–0.77], P = 7.4 × 10–7; and aHR = 1.59 [1.11–1.28], P = 0.012, respectively).
 Figure 8
Figure 8On-ART predictors of disease progression after interruption of early ART. Kaplan-Meier survival analysis for CA HIV US RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio, measured at the latest time point available before interruption of early ART, as well as detectability of CA HIV MS RNA at any time point on early ART, and time on early ART (60 or 24 weeks) (n = 51). The composite endpoint of the analysis was either a CD4+ count measurement of less than 350 cells/mm3 or restart of ART. Levels of US RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio were stratified into “high” and “low” strata based on median values. HRs and 95% CIs were calculated using Mantel-Haenszel method. P values were calculated using log-rank tests. det, detectable; ud, undetectable.
 Table 6
Table 6TI variables associated with disease progression (CD4+ count < 350 cells/mm3 or restart ART) after interruption of early ART (n = 51; Cox proportional hazards analysis)
CA HIV-1 RNA predicts disease progression in the absence of treatment. Finally, we sought to establish the predictive role of variables measured in untreated infection (at the virological set point) for the subsequent disease progression (as defined above). Participants from all 3 arms, including those who were randomized to no treatment during early ART, were included in this analysis. All virological and clinical variables measured at the set point were significantly predictive of disease progression, with high MS RNA level, low CD4+ count, and low CD4/CD8 ratio demonstrating particularly strong predictive power with hazard ratios of 0.22 (0.11–0.44), 14.48 (6.58–31.85), and 8.87 (4.20–18.75), respectively, and P value less than 0.0001 for all 3 predictors (Figure 9). Having been treated with temporary early ART was also significantly associated with longer disease progression (HR = 0.19 [0.06–0.61], P = 0.0047), confirming the earlier results of Grijsen et al. (28). HIV-1 subtype B was marginally associated with faster disease progression (P = 0.053) (Supplemental Table 7). These associations were confirmed by the univariable Cox regression analysis because all virological and clinical variables remained significantly associated with disease progression (Table 7). However, only CD4+ count and MS RNA were retained as independent predictors in the final multivariable Cox model (aHR = 0.55 [0.42–0.72], P = 1.1 × 10–5; and aHR = 1.46 [1.07–2.01], P = 0.019, respectively). The same results were obtained when the drop of CD4+ count below 350 cells/mm3 was used as the sole endpoint, censoring the participants who (re)started ART with higher CD4+ counts (data not shown).
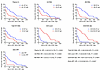 Figure 9
Figure 9Predictors of disease progression in the absence of treatment. Kaplan-Meier survival analysis for plasma VL, CA HIV US and MS RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio, measured at the virological set point (36 weeks after interruption of early ART or randomization), as well as having been treated with early ART or not (n = 56). The composite endpoint of the analysis was either a CD4+ count measurement of less than 350 cells/mm3 or (re)start of ART. Levels of plasma VL, US RNA, MS RNA, total HIV DNA, CD4+ count, and CD4/CD8 ratio were stratified into “high” and “low” strata based on median values. HRs and 95% CIs were calculated using Mantel-Haenszel method. P values were calculated using log-rank tests.
 Table 7
Table 7Set point variables associated with the rates of disease progression (time to CD4+ count < 350 cells/mm3 or restart ART) after virological set point (n = 56; Cox proportional hazards analysis)
-
Discussion
In this study, we have demonstrated substantial predictive value of CA HIV-1 RNA for (a) the virological and immunological response to early ART, (b) the magnitude of and time to viral rebound after early ART discontinuation, and (c) the subsequent disease progression in the absence of treatment. While US RNA was predictive of virological endpoints, such as time to virological suppression on ART, as well as the magnitude of and time to viral rebound after TI, MS RNA was not predictive of these endpoints. Instead, MS RNA predicted immunological endpoints: immunological response to ART and CD4+ cell loss in the absence of treatment. US RNA+ cells greatly outnumber MS RNA+ cells in both untreated and treated HIV-1 infection (refs. 33–35; our unpublished observations). This might be due to either posttranscriptional latency mechanisms (33, 36) or to (Tat-independent) transcription from defective proviruses with intact gag sequences but with deletions or other defects in the tat/rev region, splice sites, of exonic splicing enhancers. Moreover, MS RNA decays much faster than US RNA upon ART initiation (37–39). Therefore, we can speculate that while the US RNA level reflects the total number of cells harboring transcription-competent (but not necessarily replication-competent) HIV-1 (40, 41), MS RNA might mark a productively infected subset of these cells with a particularly high level of viral protein and virus production. This subset likely plays a crucial role in HIV-1 pathogenesis, defining the rates of CD4+ cell loss in the absence of therapy and CD4/CD8 ratio reconstitution on ART. Indeed, as early as 1994, in the setting of untreated HIV-1 infection, an increase in MS RNA level in PBMCs was reported to precede the drop in CD4+ count (42), and an independent predictive value of this marker for the disease progression was demonstrated a year later (43). At approximately the same time, plasma VL, which was not measured in the aforementioned studies, was shown by another group to predict disease progression independent of CD4+ count (7, 8, 44). These findings were confirmed by multiple independent studies, and plasma VL subsequently became the sole virological marker used in the clinic for more than 20 years for measuring HIV-1 replication and monitoring the response to ART. In contrast, CA RNA measurements remained confined to the research domain (41, 45). To date, this marker has not been developed for clinical use, and its associations with clinical endpoints have not been compared with other HIV-1 markers.
To the best of our knowledge, this is the first study to reveal the predictive power of CA HIV-1 RNA for prediction of a number of clinical endpoints (virological and immunological response to ART, magnitude of viral rebound, and rate of CD4+ T cell decline after TI). Moreover, this is the first study to directly compare CA HIV-1 RNA and plasma VL for prediction of a number of endpoints, including response to ART, magnitude of viral rebound, and disease progression in the absence of treatment. Remarkably, when adjusted for CA RNA, plasma VL no longer appeared as an independent predictor of any clinical endpoint in this cohort. One possible reason plasma VL was outperformed by CA RNA is the fact that plasma VL reflects a balance between virus production and virus clearance. Although clearance of free virus in HIV-1 infection was shown to be very rapid, with a half-life in the order of minutes to hours (46, 47), it may not be a simple first-order reaction, and its rate may depend on plasma VL, as shown for hepatitis B virus (48). Virus-producing cells are also cleared by host immunity (e.g., ref. 49) and die as a result of infection and their activated status, but they still have a much longer life span than free plasma virions (46). Therefore, CA HIV-1 RNA level might be a more direct measure of virus production and therefore better reflects the viral replication rate and impact on pathogenesis.
Total HIV-1 DNA, shown in some studies to be independently predictive of viral rebound and disease progression (10, 12, 25), was also outperformed by CA RNA in our study, reflecting the facts that many proviruses are replication defective and some are even transcription defective (50–52). It is therefore not surprising that the CA RNA level, which reflects the relative number of transcription-competent proviruses, appeared as a more meaningful marker than total DNA for the association with a number of clinical endpoints. One puzzling observation made in this study was that while pre-ART US RNA was positively predictive of the magnitude of viral rebound after early ART discontinuation, total DNA was negatively predictive. This latter effect was revealed only when the US RNA level was adjusted for in the multivariable analysis, providing a possible explanation of why it has not been observed previously. This indicates that pre-ART total HIV-1 DNA has an additional negative effect on viral rebound, which is independent of the positive, US RNA production–mediated, effect. The biology behind this additional effect is presently unclear, but HIV-1 DNA load in acute/early untreated infection increases with time (53–57), and thus pre-ART HIV-1 DNA in early infection should positively correlate with the degree of development of antiviral immune response at the moment of ART initiation. The latter would negatively influence the magnitude of viral rebound upon discontinuation of early ART (58–60), providing a theoretical explanation for this apparently paradoxical effect. Unfortunately, we could not test this hypothesis in our cohort because the exact infection dates were not available and the vast majority of participants were diagnosed during Fiebig stages III–IV.
Arguably the most important finding of this study is the demonstration of the predictive value of US RNA for the time to viral rebound upon TI. Validated predictive biomarkers for the time to viral rebound and posttreatment HIV-1 control are urgently needed (24), and several studies investigated such candidate predictive biomarkers. In 2008, Lewin et al. reported that plasma HIV-1 RNA at 12 weeks of ART initiated during primary infection was associated with viral rebound after structured treatment interruptions (61). In the present study, no variable measured at 12 weeks of ART, including plasma VL and CA HIV RNA and DNA, was significantly associated with time to viral rebound (data not shown). It must be noted that Lewin et al. defined a plasma VL more than 5000 copies/mL as viral rebound, whereas lower thresholds of 50 and 400 copies/mL were used in our study. In addition, Lewin et al. defined a binary outcome (maintenance of HIV plasma VL below 5000 copies/mL for 24 weeks after TI), whereas we used survival analysis. More recently, Williams et al. identified total HIV-1 DNA as predictive of disease progression and time to viral rebound after temporary early ART interruption in the Short Pulse Anti-Retroviral Therapy at Seroconversion (SPARTAC) study (25), although this was not confirmed in the subset of African participants of SPARTAC (62), and CA RNA was not measured in these studies. Interestingly, a recent study has identified total but not intact HIV-1 DNA as a marker that distinguishes posttreatment controllers from noncontrollers (63), indicating the clinical significance of defective HIV-1 proviruses. However, Li et al., who investigated participants from 6 pooled AIDS Clinical Trials Group ATI studies, did not find any significant predictive value of total HIV-1 DNA for the time to viral rebound (26). Instead, lower CA US HIV-1 RNA level and NNRTI-based ART regimen were found to be associated with longer time to rebound in 2-covariate discrete-time logistic Cox models. However, no continuous time-to-event analysis to explain time to rebound was used in that study (26). A weak but significant reverse correlation was also noted between the level of CA HIV-1 RNA (but not HIV-1 DNA) and the timing of viral rebound in a therapeutic vaccination study in HIV-infected individuals who initiated ART during early infection (64). In contrast, in the REDUC Part B trial, which analyzed the effects of a therapeutic HIV-1 vaccine followed by a latency-reversing agent romidepsin, a strong negative correlation was observed between HIV-1 DNA, and to a lesser extent CA RNA, measured approximately 2 months before ATI, and the time to viral rebound (65). The latter 2 studies also did not use time-to-event analysis to explain time to rebound. In addition, immune checkpoint molecules, such as PD-1, Tim-3, and Lag-3, have been shown to predict time to rebound (27). It must be noted that these studies used different thresholds for viral rebound, measured variables at different time points, and performed different types of statistical analysis. For example, Williams et al. found a predictive value of HIV-1 DNA for viral rebound to more than 400 but not to more than 50 HIV-1 RNA copies/mL (25), whereas only thresholds of 200 and 1000 copies/mL were used by Li et al. (26) and only the threshold of 400 copies/mL was used by Hurst et al. (27). Significant predictive value of HIV-1 DNA for the rebound to more than 50 but not to more than 1000 copies/mL was found by Moron-Lopez et al., whereas the opposite pattern was found for CA RNA (65). In addition, some studies included participants from the intervention trials (64, 65), which could have influenced the results. In summary, several candidate biomarkers have been identified, but different studies provided heterogeneous and sometimes contradictory results.
In this study, which included well-documented participants from a randomized trial, we observed a significant (Table 5) independent predictive value of CA US RNA for the time to viral rebound of both more than 50 and more than 400 copies/mL, as demonstrated by both Kaplan-Meier and Cox regression analyses that included a number of demographic and clinical covariates. This confirms the results of the previous studies that found a predictive value of US RNA for the time to viral rebound (26, 64, 65), but we believe our study design and analysis are more robust. We have previously reported that US RNA on ART predicts virological failure and correlates with small changes in therapy adherence (2, 3). Together with these earlier studies, our present results reinforce US RNA as a marker of residual HIV-1 activity on ART. Recent studies demonstrated that defective proviruses can be transcribed and that the transcription competence of a provirus does not necessarily mean replication competence (66, 67). However, the predictive power of CA RNA for a number of outcomes, including time to viral rebound, as reported here, still argues that despite being partially composed of defective proviruses, the transcription-competent reservoir does reflect the replication-competent reservoir.
No other marker, including HIV-1 DNA or the therapy regimen, was shown to be predictive for time to viral rebound in our study. One surprising observation was that longer duration of ART (60 vs. 24 weeks) did not predict longer time to rebound, contradicting the previous studies (68, 69). It is possible that the difference between 24 and 60 weeks of ART is insufficient to observe an effect on the time to rebound. It has also been shown that time from seroconversion to the start of ART is negatively associated with time to rebound and CD4/CD8 ratio normalization (68, 70). In this cohort, no difference for time to rebound was observed between participants who were diagnosed during Fiebig stages I–II versus III–IV versus V–VI. However, the vast majority of participants were diagnosed at stages III or IV, and treatment started several weeks after diagnosis (28). Therefore, exact times from infection or seroconversion to the start of ART could not be determined in our study. However, a recent study has shown that rapid viral rebound upon ATI occurs even in individuals who start treatment in Fiebig stage I of acute infection (16). It is possible that Primo-SHM participants, who were treated during PHI but most of whom did not start treatment very early in infection, had an advantage of having more time to develop anti-HIV immune responses before the start of ART. This, coupled to the potent 4-drug ART regimen that resulted in a prominent reduction in the HIV-1 reservoir size (our unpublished observations), might be one of the reasons some participants demonstrated various degrees of posttreatment control (this study and unpublished data). Another limitation of the study is that the Primo-SHM trial was not designed to measure the exact duration of posttreatment control, and therefore plasma VL was not sampled very frequently, especially starting from 3 months after TI. The study was also underpowered to detect effects of sex, transmission route, or HIV-1 subtype because the vast majority of participants were Dutch MSM infected with HIV-1 subtype B. In general, the results of this study might be applicable only to populations similar to those described in this report. The final limitation is that, because of limited PBMC sample availability, there was some variation between participants with regard to the exact time points of the measurements (Supplemental Methods). This does not compromise the validity of the main results of our study: that CA RNA measured on ART predicts time to viral rebound after TI and that CA RNA measured at untreated infection predicts disease progression.
In summary, this study positions CA HIV-1 RNA in PBMCs as a promising HIV-1 biomarker with a stronger predictive value than plasma VL for a number of clinical endpoints. We realize the technical challenges of processing blood cells compared with cell-free plasma, including the need for an internal normalizer for input cellular amounts. Therefore, we do not envision CA RNA entering routine clinical use in the near future. However, some applications demand the best assays available; therefore, further research into the potential of CA RNA as an HIV-1 clinical marker is warranted. In particular, its predictive value for viral rebound after TI should be explored further in the context of HIV-1 cure research.
-
Methods
Study population and treatment. The Primo-SHM trial was a multicenter randomized controlled trial comparing temporary early ART with no treatment during PHI. Participants were randomly assigned to receive no treatment or 24 or 60 weeks of ART (3-way randomization). If treatment was clinically indicated based on severe clinical symptoms or the participant insisted on starting early ART, subjects were randomized over the 2 treatment arms (2-way randomization). Details of trial design are reported elsewhere (28). Of 64 participants included in the present study, 50 were 3-way and 14 were 2-way randomized. Participants were recruited from May 2003 until March 2010, and follow-up data were collected until April 2015. Participants with missing values on 1 or more variables (mainly because of limited PBMC sample availability at baseline and at the virological set point) were excluded from the corresponding analyses: 8 were excluded from the baseline analysis and the same number from the set point analysis. One participant did not interrupt early ART and was excluded from the TI analysis.
Among participants, 81.3% were diagnosed with HIV-1 during Fiebig stage III or IV of early infection, and treatment started on average 4 weeks after diagnosis (28). Early ART consisted of a 4-drug, triple-class regimen: zidovudine/lamivudine (300/150 mg bid), efavirenz (600 mg qd), and lopinavir/ritonavir capsules (533/133 mg bid). After January 2008, zidovudine/lamivudine was replaced by tenofovir/emtricitabine (245/200 mg qd; Dutch standard of care). Changes to the regimen were allowed in the case of transmitted drug resistance or if one of the drugs was contraindicated or not tolerated. Of 52 participants who were randomized to one of the treatment arms, 88.5% started on triple-class therapy. Treatment was changed to a double-class, 3-drug regimen once plasma VL became undetectable. At TI, 64.7% were treated with an NNRTI-based, 25.5% with a PI-based, and 9.8% with an NNRTI + PI–based ART. At the start of early ART, the NRTI backbone consisted of zidovudine/lamivudine (63.5%) or tenofovir/emtricitabine (36.5%), and by the time of TI, it consisted of tenofovir/emtricitabine (52.9%), zidovudine/lamivudine (31.4%), or tenofovir/lamivudine (15.7%).
Measurements. Plasma VL was measured using commercial assays with detection limits of 50 or 40 copies/mL. For CA HIV-1 RNA and total HIV-1 DNA measurements, total nucleic acids were extracted from PBMCs using the Boom isolation method (71). Extracted cellular RNA was treated with DNase (DNA-free kit; Thermo Fisher Scientific) to remove DNA that could interfere with the quantitation and reverse-transcribed using random primers and SuperScript III reverse transcriptase (all from Thermo Fisher Scientific). CA HIV-1 RNA and total HIV-1 DNA were measured using previously described quantitative PCR–based (qPCR-based) methods (72, 73). HIV-1 DNA or RNA copy numbers were determined using a 7-point standard curve with a linear range of more than 5 orders of magnitude that was included in every qPCR run and normalized to the total cellular DNA (by measurement of β-actin DNA) or RNA (by measurement of 18S ribosomal RNA) inputs, respectively, as described previously (3). Nontemplate control wells were included in every qPCR run and were consistently negative.
Total HIV-1 DNA was detectable in 94.6% of participants at baseline, 82.4% at TI, and 98.3% at the virological set point; US RNA in 98.2% at baseline, 62.8% at TI, and 94.7% at the set point; and MS RNA in 70.9% at baseline, 15.7% at any visit under ART, and 67.9% at the set point. Undetectable measurements of CA RNA or DNA were assigned the values corresponding to 50% of the corresponding assay detection limits, with a maximum of 100 copies/μg total RNA or 100 copies/106 PBMCs, respectively. The detection limits depended on the amounts of the normalizer (input cellular DNA or RNA) and therefore differed among samples. Measurements with low input cellular DNA or RNA and undetectable HIV-1 DNA or RNA, respectively, were excluded from the analysis. MS RNA under ART was included in the analysis of time to viral rebound as a categorical variable (detectable vs. undetectable).
Statistics. Strength of the correlations between variables per time point and between time points per variable was assessed by Spearman’s tests. Associations of variables with the magnitude of viral rebound was assessed by fitting uni- and multivariable generalized linear models. Predictive power of variables for the clinical endpoints (response to ART, time to viral rebound after TI, CD4+ T cell decline in untreated infection) was initially assessed by Kaplan-Meier analyses with log-rank tests, nonparametric Mann-Whitney U tests, or Fisher’s exact tests, as appropriate. Subsequently, uni- and multivariable Cox proportional hazards regression models or logistic regression models were fitted. Explanatory variables that were associated with the dependent variable (P < 0.1) in univariable Cox or logistic regression analyses were included in the multivariable models, which were fitted using forward stepwise regression, except where indicated. Stepwise regression was chosen to apply an unbiased approach and at the same time to avoid model overfitting. Of note, fitting the models using backward stepwise regression did not change the outcomes. Individual tests are described in the legends to figures and tables. Data were analyzed using Prism 7.0 (GraphPad Software) and IBM SPSS Statistics (version 25). All tests were 2 sided. P values less than 0.05 were considered statistically significant.
Study approval. The study was approved by the AMC Medical Ethics Committee, and written informed consent was obtained from all participants.
-
Author contributions
AOP, MLG, FWW, JMP, and BB conceived and designed the experiments. AOP performed the experiments and analyzed the data. MB and SJ contributed reagents, materials, and analysis tools. AOP, MLG, FWW, JMP, and BB wrote the paper.
-
Acknowledgments
We are thankful to Luuk Gras and Colette Smit for their help with data collection. We would like to thank the Primo-SHM study group and study participants for helping establish this cohort. This study was supported by Aidsfonds Netherlands under grant number 2011020 to AOP.
Address correspondence to: Alexander O. Pasternak, AMC, Room K3-113B, Meibergdreef 15, 1105 AZ Amsterdam, Netherlands. Phone: 31.20.5668572; Email: a.o.pasternak@amsterdamumc.nl.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(6):e134196.https://doi.org/10.1172/jci.insight.134196.
-
References
- Deeks SG, et al. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med. 2016;22(8):839–850.
- Pasternak AO, et al. Modest nonadherence to antiretroviral therapy promotes residual HIV-1 replication in the absence of virological rebound in plasma. J Infect Dis. 2012;206(9):1443–1452.
- Pasternak AO, Jurriaans S, Bakker M, Prins JM, Berkhout B, Lukashov VV. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4(12):e8490.
- European Pregnancy Paediatric HIV Cohort Collaboration (EPPICC) Early-treated Perinatally HIV-infected Individuals: Improving Children’s Actual Life with Novel Immunotherapeutic Strategies (EPIICAL) study groups. Predictors of faster virological suppression in early treated infants with perinatal HIV from Europe and Thailand. AIDS. 2019;33(7):1155–1165.
- Ceccherini-Silberstein F, et al. Pre-ART HIV-1 DNA in CD4+ T cells correlates with baseline viro-immunological status and outcome in patients under first-line ART. J Antimicrob Chemother. 2018;73(12):3460–3470.View this article via: PubMed Google Scholar
- Tanner Z, et al. Predictors of viral suppression and rebound among HIV-positive men who have sex with men in a large multi-site Canadian cohort. BMC Infect Dis. 2016;16(1):590.
- Mellors JW, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126(12):946–954.
- Mellors JW, Rinaldo CR, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272(5265):1167–1170.
- Eller MA, et al. HIV type 1 disease progression to aids and death in a rural ugandan cohort is primarily dependent on viral load despite variable subtype and T-cell immune activation levels. J Infect Dis. 2015;211(10):1574–1584.
- Rouzioux C, et al. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis. 2005;192(1):46–55.
- Kostrikis LG, et al. Quantitation of human immunodeficiency virus type 1 DNA forms with the second template switch in peripheral blood cells predicts disease progression independently of plasma RNA load. J Virol. 2002;76(20):10099–10108.
- Goujard C, et al. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis. 2006;42(5):709–715.
- Tierney C, et al. Prognostic value of baseline human immunodeficiency virus type 1 DNA measurement for disease progression in patients receiving nucleoside therapy. J Infect Dis. 2003;187(1):144–148.
- Henrich TJ, et al. HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: An observational study. PLoS Med. 2017;14(11):e1002417.
- Chun TW, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010;24(18):2803–2808.
- Colby DJ, et al. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med. 2018;24(7):923–926.
- Pasternak AO, Berkhout B. HIV reservoir: finding the right needles in a needlestack. Cell Host Microbe. 2016;20(3):280–282.
- Sharaf RR, Li JZ. The alphabet soup of HIV reservoir markers. Curr HIV/AIDS Rep. 2017;14(2):72–81.
- Strategies for Management of Antiretroviral Therapy (SMART) Study Group, et al. CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med. 2006;355(22):2283–2296.
- Julg B, et al. Recommendations for analytical antiretroviral treatment interruptions in HIV research trials-report of a consensus meeting. Lancet HIV. 2019;6(4):e259–e268.
- Sáez-Cirión A, et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 2013;9(3):e1003211.
- Van Gulck E, et al. Immune and viral correlates of “secondary viral control” after treatment interruption in chronically HIV-1 infected patients. PLoS One. 2012;7(5):e37792.
- Martin GE, Frater J. Post-treatment and spontaneous HIV control. Curr Opin HIV AIDS. 2018;13(5):402–407.
- Li JZ, Smith DM, Mellors JW. The need for treatment interruption studies and biomarker identification in the search for an HIV cure. AIDS. 2015;29(12):1429–1432.
- Williams JP, et al. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife. 2014;3:e03821. View this article via: PubMed Google Scholar
- Li JZ, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS. 2016;30(3):343–353.View this article via: PubMed Google Scholar
- Hurst J, et al. Immunological biomarkers predict HIV-1 viral rebound after treatment interruption. Nat Commun. 2015;6:8495. View this article via: PubMed Google Scholar
- Grijsen ML, et al. No treatment versus 24 or 60 weeks of antiretroviral treatment during primary HIV infection: the randomized Primo-SHM trial. PLoS Med. 2012;9(3):e1001196.
- Steingrover R, et al. Transient lowering of the viral set point after temporary antiretroviral therapy of primary HIV type 1 infection. AIDS Res Hum Retroviruses. 2010;26(4):379–387.
- Desquilbet L, et al. Does transient HAART during primary HIV-1 infection lower the virological set-point? AIDS. 2004;18(18):2361–2369.View this article via: PubMed Google Scholar
- Serrano-Villar S, et al. HIV-infected individuals with low CD4/CD8 ratio despite effective antiretroviral therapy exhibit altered T cell subsets, heightened CD8+ T cell activation, and increased risk of non-AIDS morbidity and mortality. PLoS Pathog. 2014;10(5):e1004078.
- Han WM, et al. CD4/CD8 ratio normalization rates and low ratio as prognostic marker for non-AIDS defining events among long-term virologically suppressed people living with HIV. AIDS Res Ther. 2018;15(1):13.
- Yukl SA, et al. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med. 2018;10(430):eaap9927.
- Kaiser P, Joos B, Niederöst B, Weber R, Günthard HF, Fischer M. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol. 2007;81(18):9693–9706.
- Massanella M, et al. Improved assays to measure and characterize the inducible HIV reservoir. EBioMedicine. 2018;36:113–121.
- Sarracino A, et al. Posttranscriptional regulation of HIV-1 gene expression during replication and reactivation from latency by nuclear matrix protein MATR3. mBio. 2018;9(6):e02158-18. View this article via: PubMed Google Scholar
- Bagnarelli P, et al. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J Virol. 1996;70(11):7603–7613.
- Vesanen M, Markowitz M, Cao Y, Ho DD, Saksela K. Human immunodeficiency virus type-1 mRNA splicing pattern in infected persons is determined by the proportion of newly infected cells. Virology. 1997;236(1):104–109.
- Zanchetta M, et al. Long-term decay of the HIV-1 reservoir in HIV-1-infected children treated with highly active antiretroviral therapy. J Infect Dis. 2006;193(12):1718–1727.
- Baxter AE, O’Doherty U, Kaufmann DE. Beyond the replication-competent HIV reservoir: transcription and translation-competent reservoirs. Retrovirology. 2018;15(1):18.
- Pasternak AO, Berkhout B. What do we measure when we measure cell-associated HIV RNA. Retrovirology. 2018;15(1):13.
- Saksela K, Stevens C, Rubinstein P, Baltimore D. Human immunodeficiency virus type 1 mRNA expression in peripheral blood cells predicts disease progression independently of the numbers of CD4+ lymphocytes. Proc Natl Acad Sci U S A. 1994;91(3):1104–1108.
- Saksela K, Stevens CE, Rubinstein P, Taylor PE, Baltimore D. HIV-1 messenger RNA in peripheral blood mononuclear cells as an early marker of risk for progression to AIDS. Ann Intern Med. 1995;123(9):641–648.
- Mellors JW, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122(8):573–579.
- Pasternak AO, Lukashov VV, Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. 2013;10:41. View this article via: PubMed Google Scholar
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271(5255):1582–1586.
- Ramratnam B, et al. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet. 1999;354(9192):1782–1785.
- Dandri M, Murray JM, Lutgehetmann M, Volz T, Lohse AW, Petersen J. Virion half-life in chronic hepatitis B infection is strongly correlated with levels of viremia. Hepatology. 2008;48(4):1079–1086.
- Lu CL, et al. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science. 2016;352(6288):1001–1004.
- Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551.
- Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049.
- Wiegand A, et al. Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc Natl Acad Sci U S A. 2017;114(18):E3659–E3668.
- Jain V, et al. Antiretroviral therapy initiated within 6 months of HIV infection is associated with lower T-cell activation and smaller HIV reservoir size. J Infect Dis. 2013;208(8):1202–1211.
- Schmid A, et al. Profound depletion of HIV-1 transcription in patients initiating antiretroviral therapy during acute infection. PLoS One. 2010;5(10):e13310.
- Kuhn L, et al. Age at antiretroviral therapy initiation and cell-associated HIV-1 DNA levels in HIV-1-infected children. PLoS One. 2018;13(4):e0195514.
- Watanabe D, et al. Cellular HIV-1 DNA levels in patients receiving antiretroviral therapy strongly correlate with therapy initiation timing but not with therapy duration. BMC Infect Dis. 2011;11:146. View this article via: PubMed Google Scholar
- Ananworanich J, et al. HIV DNA set point is rapidly established in acute HIV infection and dramatically reduced by early ART. EBioMedicine. 2016;11:68–72.
- Geldmacher C, et al. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol. 2007;81(5):2440–2448.
- Zuñiga R, et al. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol. 2006;80(6):3122–3125.
- Rolland M, et al. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One. 2008;3(1):e1424.
- Lewin SR, et al. Virologic determinants of success after structured treatment interruptions of antiretrovirals in acute HIV-1 infection. J Acquir Immune Defic Syndr. 2008;47(2):140–147.View this article via: PubMed Google Scholar
- Gossez M, et al. Virological remission after antiretroviral therapy interruption in female African HIV seroconverters. AIDS. 2019;33(2):185–197.
- Sharaf R, et al. HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J Clin Invest. 2018;128(9):4074–4085.
- Sneller MC, et al. A randomized controlled safety/efficacy trial of therapeutic vaccination in HIV-infected individuals who initiated antiretroviral therapy early in infection. Sci Transl Med. 2017;9(419):eaan8848.
- Moron-Lopez S, Kim P, Søgaard OS, Tolstrup M, Wong JK, Yukl SA. Characterization of the HIV-1 transcription profile after romidepsin administration in ART-suppressed individuals. AIDS. 2019;33(3):425–431.
- Imamichi H, et al. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A. 2016;113(31):8783–8788.
- Pollack RA, et al. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe. 2017;21(4):494–506.e4.
- Fidler S, et al. Virological blips and predictors of post treatment viral control after stopping ART started in primary HIV Infection. J Acquir Immune Defic Syndr. 2017;74(2):126–133.
- Stöhr W, et al. Duration of HIV-1 viral suppression on cessation of antiretroviral therapy in primary infection correlates with time on therapy. PLoS One. 2013;8(10):e78287.
- Thornhill J, et al. Brief report: enhanced normalization of CD4/CD8 ratio with earlier antiretroviral therapy at primary HIV infection. J Acquir Immune Defic Syndr. 2016;73(1):69–73.
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28(3):495–503.
- Pasternak AO, et al. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46(7):2206–2211.
- Malnati MS, et al. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc. 2008;3(7):1240–1248.
- Fischer M, et al. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol. 1999;37(5):1260–1264.
-
Version history
- Version 1 (February 25, 2020): In-Press Preview
- Version 2 (March 26, 2020): Electronic publication
- Version 3 (March 26, 2020): The legend for Figure 9 was updated.



















