Resource and Technical AdvanceTherapeutics Free access | 10.1172/jci.insight.98305
Selection of phage-displayed accessible recombinant targeted antibodies (SPARTA): methodology and applications
Sara D’Angelo,1 Fernanda I. Staquicini,2 Fortunato Ferrara,1 Daniela I. Staquicini,2 Geetanjali Sharma,3 Christy A. Tarleton,3 Huynh Nguyen,3 Leslie A. Naranjo,1 Richard L. Sidman,4 Wadih Arap,5 Andrew R.M. Bradbury,1 and Renata Pasqualini2
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by D’Angelo, S. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Staquicini, F. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Ferrara, F. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Staquicini, D. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Sharma, G. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Tarleton, C. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Nguyen, H. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Naranjo, L. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by
Sidman, R.
in:
PubMed
|
Google Scholar
|

1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Arap, W. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Bradbury, A. in: PubMed | Google Scholar
1Specifica Inc., Santa Fe, New Mexico, USA.
2Rutgers Cancer Institute of New Jersey at University Hospital and Division of Cancer Biology, Department of Radiation Oncology, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
3University of New Mexico Comprehensive Cancer Center, University of New Mexico School of Medicine, Albuquerque, New Mexico, USA.
4Department of Neurology, Harvard Medical School, Boston, Massachusetts, USA.
5Rutgers Cancer Institute of New Jersey at University Hospital and Division of Hematology/Oncology, Department of Medicine, Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Find articles by Pasqualini, R. in: PubMed | Google Scholar
Authorship note: SD, FIS, and FF equally contributed to this work. WA, ARMB, and RP equally contributed to this work.
Published May 3, 2018 - More info
JCI Insight. 2018;3(9):e98305. https://doi.org/10.1172/jci.insight.98305.
Copyright © 2018, American Society for Clinical Investigation
Received: October 25, 2017; Accepted: April 5, 2018
-
Abstract
We developed a potentially novel and robust antibody discovery methodology, termed selection of phage-displayed accessible recombinant targeted antibodies (SPARTA). This combines an in vitro screening step of a naive human antibody library against known tumor targets, with in vivo selections based on tumor-homing capabilities of a preenriched antibody pool. This unique approach overcomes several rate-limiting challenges to generate human antibodies amenable to rapid translation into medical applications. As a proof of concept, we evaluated SPARTA on 2 well-established tumor cell surface targets, EphA5 and GRP78. We evaluated antibodies that showed tumor-targeting selectivity as a representative panel of antibody-drug conjugates (ADCs) and were highly efficacious. Our results validate a discovery platform to identify and validate monoclonal antibodies with favorable tumor-targeting attributes. This approach may also extend to other diseases with known cell surface targets and affected tissues easily isolated for in vivo selection.
-
Introduction
Monoclonal antibody–based therapy of human cancers has emerged as a major advance in contemporary medical oncology. However, the identification of suitable cancer target candidates still remains the initial step to develop clinical applications. Cancer-specific targets often localize abundantly on the surface of cancer cells or nonmalignant tumor–associated vascular endothelial and stromal cells, so they are accessible through systemic circulation (1). Such targets usually consist of a broad array of proteins that are overexpressed, mutated, or abnormally located in the cell surface compared with normal tissues (2). Conventionally, a target candidate is identified and validated, panels of antibodies are produced and evaluated for biological activity, and then favorable immune profiles are established prior to drug lead optimization. The more recent use of in vitro technologies, such as phage and yeast display, to generate monoclonal antibody clones has some advantages over traditional immunization. These include the speed with which antibody clones undergo selection and isolation, the ability to enrich specific properties in high-throughput procedures, and perhaps most importantly, direct selection of human monoclonal antibodies.
To discover target-specific, biologically active antibodies to common human cancers, we developed a 2-step, in-tandem methodology: selection of phage-displayed accessible recombinant targeted antibodies (termed SPARTA). Unlike blind selection approaches with no knowledge about the target, SPARTA begins with a previously identified tumor cell–surface target. An enriched pool of recombinant human antibodies against this target is first generated from a large naive human library in vitro using a high-throughput combination of phage and yeast displays (3). Then, antibodies from this pool are directly selected in vivo for their tumor-targeting attributes. To our knowledge, our group has pioneered the experimental use of in vivo peptide phage (1, 4–13), and we have extensively documented that this approach provides exquisite advantages to identify accessible target receptors in the unique context of the native tumor microenvironment. We and other investigators have attempted to extend the in vivo methodology to antibody phage–display libraries. However, we found only modest success (14–17), likely due to inherent technical constraints such as the critical need for helper phage rescue, which leads to extremely low antibody display levels. In addition, the presence of truncated antibodies and associated exposure of hydrophobic interfaces generally increases nonspecific binding to nearly prohibitive background levels.
As proof of concept, we examined 2 established cancer cell–surface targets, Ephrin A5 (EphA5; a molecular target in human lung cancer; ref. 18) and 78 kDa glucose-regulated protein (GRP78; a relatively promiscuous target on the tumor cell surface of several human cancers; ref. 19–21). In fact, monoclonal antibodies have been successfully generated against both human EphA5 (18) and GRP78 (22–24), albeit at high cost and low translational value. Generation of an enriched pool of human recombinant antibodies screened against EphA5 and GRP78 in vitro, along with the functional in vivo selection of monoclonal antibody pools in preclinical models of human lung cancer and breast cancer, led to the identification of single monoclonal antibody clones with favorable tumor-targeting properties. The individual monoclonal antibody clones against EphA5 and GRP78 consistently recognized and localized to their cognate tumor targets in vivo and showed effective killing activity as antibody-drug conjugates (ADCs). We conclude that SPARTA is a broad platform to select tumor-specific antibodies from large human phage–display antibody libraries. SPARTA may become the method of choice to select monoclonal antibodies against human cancers and, perhaps, certain nonmalignant diseases.
-
Results
Serial in vitro screening and in vivo selection to discover human recombinant monoclonal antibodies. We depict our schematic representation of SPARTA in Figure 1A. First, we conducted an in vitro unbiased library screening against immobilized human recombinant EphA5 and GRP78 from a large human naive phage-displayed single chain variable fragment (scFv) library (25). After 2 rounds of library screening in vitro, we reduced the diversity by 5 orders of magnitude (from ~1 × 1011 unique scFv sequences to <1 × 106). We then cloned the total phage pool output into a yeast-display system. This maneuver allowed precise fluorescence-activated cell sorting to restrict reactivity to those clones recognizing the target (26). After 2 additional rounds of yeast cell sorting, we obtained a diverse, highly enriched antibody population for each target (Figure 1, B and C). Antibody clones bound specifically to the corresponding antigens, in both yeast- and phage-display contexts (Figure 1, B and C). The anti-EphA5 antibody pools were negatively selected to minimize or eliminate anti-EphA5 antibodies that would also recognize orthologous ephrin family members (namely EphA3, -4, -6, and -7) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98305DS1). Next-generation sequencing (NGS) plus AbMining toolbox analysis (27) confirmed highly diverse antibody populations, as assessed by the sizable number of individual heavy chain complementarity-determining region 3 (HCDR3) variants obtained for EphA5 (n = 207) and GRP78 (n = 125). The observed relative high polyclonality underscores the power of the combined phage/yeast display approach to select diverse specific antibodies.
 Figure 1
Figure 1SPARTA methodology. (A) A naive human single-chain variable fragment (scFv) library (~1 × 1011 transducing units [TU]) is screened in vitro against immobilized recombinant antigens. The phage output pool (~1 × 105 TU) is subsequently transferred to a yeast display vector for 2 additional screening steps. After rounds of positive and negative sorting, the yeast output pool (~1 × 104 TU) is expressed multivalently in phage particles and administered i.v. into tumor-bearing mice (n = 3). Tumor-homing phage particles are recovered, amplified by PCR, and reexpressed in multivalent format for 2 additional rounds of in vivo selection. After next-generation sequencing (NGS), clonal diversity and ranking are determined. (B and C) Flow cytometry profiles and ELISA. Positive binders are shown for selected anti-EphA5 or anti-GRP78. Each dot in the FACS plot represents an individual yeast antibody–displaying clone. ELISA performed with the corresponding recombinant antigen confirmed binding specificity. Open circles represent individual data points (n = 3). (D and E) Following in vitro screening steps, 3 rounds of in vivo selection occurred in mice bearing human lung cancer xenografts for EphA5 or isogenic mammary tumors for GRP78. Open circles represent individual data points (n = 3). Tumors 1 and 2 represent 2 independent experiments. Data represent ± SEM.
Based on the current understanding of the importance of multivalency in in vivo peptide phage display originally developed by our group (1, 4, 5, 7–11, 13, 28–33), we generated multivalent antibody phage displaying anti-EphA5 and anti-GRP78 scFv populations using our helper plasmid technology (34). As opposed to traditional helper phage–mediated monovalent antibody display, helper plasmids provide all the required viral packaging functions, but because they lack the phage packaging signal, helper phage contamination is eliminated. Furthermore, different plasmids allow packaging in mono- or multivalent forms, potentially reducing background binding during in vivo selections and increasing specific binding of targeted phage through improved avidity (34). While it is somewhat challenging to use helper plasmids with large naive antibody libraries, applications to peptide and small targeted antibody libraries are much more successful (35). We cloned recombinant scFv genes previously sorted by yeast display back into the phagemid display vector pDAN5 (25), as a polyclonal pool, and transformed this into E. coli carrying the M13cp-dg3 helper plasmid (34, 35). We then administered the polyclonal multivalent phage pools i.v. into breast tumor– or lung tumor–bearing mice. We collected tumor xenografts (NCI-H460 human lung cancer cells for EphA5) or isogenic tumors (Ef43.fgf4 murine breast cancer cells for GRP78), as well as control organs, after 3 hours to enrich for phage clones that exclusively localized to tumors in vivo (Figure 1, D and E). We assessed the relative number of phage particles in tumor and control tissue samples by quantitative PCR (qPCR). Then, amplified scFv genes isolated from the tumors were recloned back into the original phage-display vector for use in serial rounds of selection in vivo. After 3 rounds, we observed specific targeting of EphA5- (Figure 1D) and GRP78-expressing tumors (Figure 1E) compared with several negative control organs. A complete list of control organs (both positive and negative) is shown in Supplemental Figure 2. We used 2 mutually nonexclusive approaches to identify lead monoclonal antibody candidates. For GRP78-targeting antibodies, we evaluated phage clones from the final round of in vivo selection for binding to the cognate recombinant antigen. For EphA5-targeting antibodies, we assessed the enrichment of tumor-homing clones by NGS and ranked clones by frequency to chose lead monoclonal antibody candidates. We isolated several different antibodies for each target that homed specifically and localized to the corresponding tumor using either of these approaches. In both cases, we verified monoclonal antibodies by DNA sequencing and confirmed full-length scFvs with no rearrangements, stop codons, or frameshifts.
Validation of tumor homing in vivo. We selected a total of 4 scFv clones against EphA5 (termed E1–4) and 3 against GRP78 (termed G1–3) for further functional studies (Supplemental Table 1). We displayed monoclonal antibodies multivalently on phage and individually tested their ability to home and localize to tumors in vivo after i.v. administration. Phage displaying a scFv antibody recognizing M2 (36), a conserved influenza virus protein, served as a standard negative control. We administered individual phage clones i.v. in tumor-bearing mice and then harvested tumor tissue and control organs after 3 hours (Figures 2 and 3). Relative quantification of phage particles in tissue samples revealed marked accumulation of EphA5-binding phage in EphA5-expressing tumors (Figure 2, A–D) and GRP78-binding phage in GRP78-expressing tumors (Figure 3, A–C) compared with several negative control organs (shown are brain, muscle, and pancreas). Additional control organs are shown in Supplemental Figures 3 and 4. We detected no tumor homing by control phage (insertless phage and phage displaying a scFv antibody recognizing M2) in either experimental tumor model.
 Figure 2
Figure 2Anti-EphA5 monoclonal antibodies home to human lung cancer xenografts in vivo. (A–D) Selected anti-EphA5 antibody clones (termed E1–4) target EphA5-expressing tumors in a xenograft model of lung cancer. A representative graph indicates each targeted phage clone. Phage displaying an antibody fragment against the viral protein M2 serves as negative control. Experiments were replicated at least twice. A representative experiment is shown. Open circles represent individual data points (n = 3). Data represent ± SEM. *P < 0.05 (Student’s t test) (tumor vs. control organs).
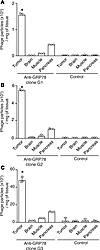 Figure 3
Figure 3Anti-GRP78 monoclonal antibodies home to isogenic breast tumors in vivo. (A–C) Selected anti-GRP78 antibody clones (termed G1–3) target an isogenic model of mammary cancer–expressing cell surface GRP78. A representative graph shows each targeted phage clone. Phage displaying an antibody fragment against the viral protein M2 serves as a negative control. Experiments were replicated at least twice. A representative experiment is shown. Open circles represent individual data points (n = 3). Data represent ± SEM. *P < 0.05 (Student’s t test)(tumor vs. control organs).
Assessment of functional binding specificity. Having validated tumor homing in vivo, we next evaluated binding specificity to recombinant proteins and endogenous corresponding targets expressed on the tumor cell surface. All individual monoclonal antibodies in multivalent phage-display format bound to their respective antigens as measured by recombinant protein–based ELISA (Figure 4, A and B) and cell-based ELISA (Figure 4, C and D). We showed receptor-mediated cell internalization of targeted phage clones upon incubation of cells expressing surface EphA5 (Figure 4E) or GRP78 (Figure 4F) with respective scFv phage, followed by stripping any residual surface-bound phage. By using different GRP78-displaying cell lines throughout this work, we demonstrated the versatility of the SPARTA method in various settings and determined the best cell line for each assay. For instance, the MCF7 cell line has better adhesion properties than Ef43.fgf4, so it was optimal for the internalization experiments that involved harsh washing conditions. Negative controls included an insertless phage, phage displaying an unrelated scFv, and control cells. In order to test if antibodies were functional proteins, we cloned scFvs into our previously described scFv-human-Fc expression vectors (37) produced in either CHO cell culture supernatants or the S. cerevisiae expression system, as indicated. Anti–GRP78 scFv-Fcs recognized human GRP78 on ELISA and flow cytometry (Supplemental Figure 5). Similarly, anti–EphA5 scFv-Fcs bound to immobilized recombinant EphA5 and endogenous EphA5 present on the cell surface (Supplemental Figure 6). When evaluated in vivo, the scFv-Fcs infiltrated and localized to tumors after a 6-minute circulation time, as assessed by immunofluorescence (IF) (Supplemental Figure 7). Taken together, these results indicate that the combination of in vitro screenings on recombinant cell surface receptors followed by in vivo selection of receptor-specific antibodies yields antibody clones with favorable on-target biodistribution.
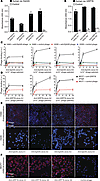 Figure 4
Figure 4Characterization of scFv-displaying phage in vitro. (A and B) Equimolar amounts of selected phage displaying anti-EphA5 clones or anti-GRP78 clones tested individually for binding to recombinant antigens. Negative controls included an anti-M2 clone and BSA. Open circles represent individual data points. Data represent ± SEM. (C and D) Binding to antigens expressed on the cell surface tested by standard phage-ELISA on cells. (C) EphA5-positive (H460) and EphA5-negative (H226) cells grown in 96-well plates were exposed to increasing concentrations of phage displaying anti-EphA5 or control scFvs. All 4 monoclonal scFv antibodies bound specifically to H460 lung cancer cells. (D) Similarly, all anti-GRP78 clones bound to GRP78-expressing breast cancer cells (MCF7), whereas a control phage showed only background binding. (E and F) Receptor-mediated internalization of anti-EphA5 and anti-GRP78 clones was assessed by immunofluorescence, after stripping of residual surface bound clones. (E) Internalization of anti-EphA5 phage clones was only observed in EphA5-expressing cells (H460), but not in EphA5-negative cells (H226) (400× magnification). (F) Anti-GRP78 clones internalized specifically in GRP78-positive MCF7 human breast cancer cells. No cell internalization was observed when using a negative control phage (400× magnification).
Monoclonal antibodies have specific cytotoxicity against lung and breast cancer cells. We tested ADCs using drug-conjugated secondary reagents recognizing the Fc region to deliver cytotoxic drugs. We prioritized monoclonal antibody candidates based on binding and receptor-mediated internalization in target-expressing cells and used a cytotoxicity assay to determine the potency of leading ADC candidates in a cell-based assay. Then, we assessed binding specificity and antigen-dependent efficacy with tumor cell lines expressing variable levels of the targets on their surfaces (Supplemental Figure 8). Of note, we deemed moderate knockdown of GRP78 (less than 50%) sufficient to prevent translocation of the receptor to the cell surface. This is consistent with the generally accepted theory that GRP78 overexpression and consequent saturation of its ER receptors leads to its atypical exposure on the membrane (38, 39). Negative controls included cells exposed to the primary scFv-Fc alone, cell lines with low but still detectable levels of EphA5 and GRP78, a nonspecific primary scFv-Fc, and drug-conjugated secondary antibody alone.
We exposed tumor cells to increasing concentrations of the leading scFv-Fcs, followed by secondary reagents conjugated to a representative panel of cytotoxic drugs: α-amanitin (AAMT), monomethyl auristatin F (MMAF), duocarmycin (DMDM), and emtansine (DM1) (Figure 5). EphA5-expressing cells showed sensitivity to scFv-Fc E4, when combined with all cytotoxic drugs. In particular, we observed a clear concentration-dependent response at a low nanomolar range for both AAMT and DMDM, which supports functional retention of both receptor-binding specificity and cell internalization attributes (Figure 5, A and B). Similarly, we found anti–GRP78 scFv-Fc G1 was most efficient at inducing cell death, although at a higher nanomolar range (Figure 5, E–H). Control tumor cells were not affected under the same experimental conditions (Figure 5, A–H). Incubation with an irrelevant negative control scFv-Fc (Figure 5, A–H, blue line) or drug-conjugated secondary antibody alone (not shown) did not result in detectable tumor cell death. We provided a complete panel of ADC experiments for reference (Supplemental Figure 9).
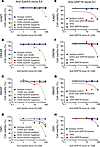 Figure 5
Figure 5Cytotoxic profile of anti-EphA5 and anti-GRP78 monoclonal antibodies. Cytotoxicity was measured in real time in the presence of anti–human IgG Fc Fab fragments (secondary antibody) conjugated to monomethyl auristatin F (MMAF) or duocarmycin (DMDM) with a cleavable linker, and either emtansine (DM1) or amanitin (AAMT) with a noncleavable linker. Optimal concentrations of the secondary antibody drug conjugate (20 nM, or 1 ng/μl) was determined as the minimal concentration not inducing significant cell toxicity. (A–D) Treatment of cells with anti–EphA5 scFv-Fc in presence of secondary antibodies conjugated to AAMT, DMDM, MMAF, and DM1 induced potent cell death of EphA5-expressing cells. An isotype control scFv-Fc in the presence of secondary antibody conjugates showed no toxicity (blue line, 100% cell viability). (E–H) GRP78-targeted scFv-Fc were more effective at killing GRP78-expressing Ef43.fgf4 cells when in the presence of AAMT- or DMDM-conjugated secondary antibody drug conjugates, whereas control GRP78-silenced Ef43.fgf4 cells were not affected. An isotype control scFv-Fc in the presence of secondary antibody conjugates showed no toxicity (blue line, 100% cell viability).
-
Discussion
The introduction of monoclonal antibodies for antibody-based cancer therapy was made possible with the development of the hybridoma technology (40). However, clinical applications of murine monoclonal antibodies were challenged by their immunogenicity and rapid clearance. Consequently, recombinant technologies and antibody humanization appeared as viable options to tentatively produce human monoclonal antibodies that would ideally be indistinguishable from those found in vivo and with the lowest expected risk of immunogenicity. This usually occurs through immunization of transgenic mice (41), humanization (42–45) of murine antibodies (40, 46–48), or in vitro display methods, particularly phage (49) and/or yeast display (50). The latter method does not require immunization and may identify human monoclonal antibodies directly from large naive human libraries (25). The combination of these 2 complementary antibody display platforms into a single selection strategy has provided 2 key potential advantages over either platform alone: screening of a large number of clones in a single experiment and tailored selection for precise functional attributes (3). Here, we introduce the SPARTA methodology, which serially integrates a 2-step strategy based on in vitro screening and in vivo selection to yield a robust monoclonal antibody discovery pipeline. Technical improvements include recloning sorted yeast-display antibodies back into our phage display vector (25), displaying them in a multivalent format (34), administering them systemically into tumor-bearing mice, and isolating individual phage clones that homed to tumors.
As proof of concept, we applied SPARTA to isolate human recombinant monoclonal antibodies against the RTK EphA5, a validated target in lung cancer (18), and the heat shock protein GRP78, a promiscuous marker of several human cancers (19, 22–24, 39, 51–54). The in vitro selections isolated hundreds of human recombinant antibodies against either EphA5 or GRP78, whereas the in vivo functional screenings in tumor-bearing mice identified those antibodies from the polyclonal pools that recognized the cell surface–associated targets within their in vivo context. Thus, SPARTA represents a significant improvement over antibody selection strategies conducted blindly, where cancer patients are infused with naive phage antibody libraries (14, 15). Although this approach successfully isolated cancer targeting antibodies, the absence of selective pressure for either specific tumor binding or reactivity with similar tumors in different patients resulted in antibodies that recognized normal tissues (14, 15) or were patient specific (14). By focusing on tumor-specific targets rather than patient-specific targets, SPARTA encourages the selection of the best therapeutic leads with the highest potential in clinical settings.
We applied a stochastic approach to GRP78-targeting selection, where a random screening of individual clones (n = 45) identified 3 different scFvs. In the case of EphA5, we carried out NGS before and after each selection step in vivo, so we could identify EphA5-binding clones highly enriched in the tumor. The effectiveness of both of these 2 mutually nonexclusive approaches demonstrates the broad utility of SPARTA, even in situations with no access to NGS and antibody analysis.
Finally, we investigated these candidates individually as phage-displayed monoclonal antibodies and demonstrated that they: (a) bind to their respective antigens as both phage and proteins, (b) bind tumor lines expressing the antigens on their cell-surface membrane, and (c) undergo receptor-mediated internalization into target-expressing tumor cells. Together, our data provide strong evidence that SPARTA produces targeted monoclonal antibodies with translational potential, since the efficacy of monoclonal antibody–based therapy relies on selective uptake by cancer cells. When produced in the scFv-Fc format, antibody fusions retained all the binding functions of the original scFvs, and ADCs demonstrated tumor cell death uniquevocally (Figure 5 and Supplemental Figure 9).
In summary, this study establishes SPARTA as a robust methodology for prompt identification of tumor-targeting human recombinant monoclonal antibodies with high specificity against established cell surface antigens. Our results show that SPARTA is well suited to become the standard to develop therapeutic antibodies from large human monoclonal antibody libraries.
-
Methods
Mice. Female WT Balb/c and female Balb/c Nu/Nu mice (6–8 weeks old) were obtained from Harlan Laboratories.
Cell culture. NCI-H460, A549, and NCI-H226 human lung cancer–derived cell lines were purchased from American Type Culture Collection (ATCC) and maintained in DMEM containing 10% FBS (Gibco) plus 1% penicillin G/streptomycin SO4 (Thermo Fisher Scientific). Human MCF7 breast cancer–derived cells were purchased from ATCC and maintained in DMEM, supplemented with 0.01 mg/ml human recombinant insulin (MilliporeSigma) and 10% FBS. Mouse mammary Ef43.fgf4 (55) cells were maintained in DMEM supplemented with 10% FBS, 5 ng/ml mouse EGF (Thermo Fisher Scientific) and 1 μg/ml bovine insulin (MilliporeSigma) plus 1% penicillin G/streptomycin SO4. SUM190PT human inflammatory breast cancer cells (BioIVT) were maintained in Ham F12 medium, supplemented with 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone (MilliporeSigma), 10 mM HEPES (Thermo Fisher Scientific), and 1% penicillin G/streptomycin SO4. CHO-S cells were purchased from Invitrogen and maintained in serum-free FreeStyle CHO Expression Medium (Gibco). Ef43.fgf4 GRP78-knockdown cells were obtained by lentivirus infection, stably transfected, and maintained in selection media containing antibiotics. The Quantum Simply Cellular Microspheres (Bangs Laboratories) were used to quantitate the number of targeted molecules on the surface of cells by flow cytometry.
Phage- and yeast-display antibody screening in vitro. scFv antibody clones were isolated by integrating phage- and yeast-display methodologies as described (26). Briefly, a naive human phage antibody library (25) was used in 2 rounds of selection in vitro either on recombinant human EphA5 (R&D Systems) or GRP78 (Abcam). Streptavidin-conjugated magnetic beads (Dynabeads) were coated with biotinylated EphA5 or GRP78, washed, and incubated with the antibody phage library. After removal of unbound phage, the remaining phage particles were recovered from the beads by acid elution and used to infect F’ pilus-carrying bacteria (Ominmax-2T1, Thermo Fisher Scientific). The isolated phage were subsequently propagated, and the cycle was reiterated. After the second round of selection, the pool of scFv clones were PCR amplified and subcloned into the pDNL6 yeast display vector (56). In this vector, the scFv gene is expressed as C-terminal fusion to the Aga2 protein of S. cerevisiae for surface display. The yeast minilibraries were further enriched for target-specific binders by applying 2 rounds of standard flow cytometry sorting (FACSAria, Becton Dickinson), as described (3, 57). Up to 10,000 individual yeast cells with positive antigen binding and scFv display were sorted and propagated for further rounds of selection.
Experimental tumor xenograft and isogenic models. Mouse mammary Ef43.fgf4 isogenic tumor cells and SUM190PT human inflammatory breast cancer cells were collected at 70% confluency and administered s.c. in the mammary fat pad of either immunocompetent female Balb/c mice (Ef43.fgf4) or female Balb/c nude mice (SUM190PT) as indicated. SUM190PT cells were administered s.c. in 1:1 (vol/vol) with Matrigel (Corning), as described (20). NCI-H460 human lung cancer cells were administered s.c. in the right flanks of Balb/c female nude mice. After approximately 10 days, tumors reached about 200–300 mm3 and were separated into size-matched tumor-bearing mouse cohorts for experimentation.
Phage display in vivo. In vivo phage selections were performed as described (8–10, 13). Animals received 1 × 1010 phage particles i.v., and entire tumors and control organs were collected after 3 hours of systemic circulation. Tumor-homing phage were retrieved by PCR amplification, and full-length scFv products (~800 bp) were recloned into the pDAN5 phagemid vector (25) and transformed into the DG3 Helper cell system described in ref. 34 to produce multivalent functional phage particles for subsequent rounds of selection. Phage quantification was performed by quantitative phage PCR (30) and host bacterial infection as described (8, 10, 11, 13).
Phage binding assays and ELISA. Serial dilutions of individual phage particles in PBS containing 2% nonfat milk (Bio-Rad) were placed in microtiter wells previously coated with 0.5 μg of either EphA5 or GRP78, as indicated. After extensive washes, remaining bound phage particles were detected with an anti-M13 peroxidase–conjugated mouse monoclonal antibody (GE Healthcare Life Sciences, catalog 27-9421-01).
Phage binding to the surface of cells was tested by whole-cell ELISA. Briefly, exponentially growing cells were fixed in 96-well microtitration plates (Nunc) at 3 × 105 cells/well and exposed to serial dilutions of either targeted or control phage particles for 2 hours at room temperature (RT). Wells were extensively washed with PBS containing 0.1% Tween-20 (MilliporeSigma), and bound particles were detected with an anti-M13 peroxidase–conjugated mouse monoclonal antibody. A nontargeted helper phage (termed M13KO7; ref. 3) and unrelated scFv served as negative controls, as indicated.
Cell internalization assay. Cell internalization assays were performed as described (19). In brief, cells plated in 8-chamber slides (Thermo Fisher Scientific) were blocked with DMEM containing 2% FBS for 1 hour at RT and incubated with 1 × 109 TU of phage. After 2 hours of incubation at 37°C, cell membrane–bound phage were removed by washes with 20 mM glycine (MilliporeSigma, pH 2.3) and fixed with PBS containing 4% paraformaldehyde (PFA, Thermo Fisher Scientific). Fixed cells were permeabilized with PBS containing 0.1% Triton X-100 (MilliporeSigma), blocked with PBS containing 1% BSA (MilliporeSigma), and incubated with a mouse anti-M13 phage monoclonal antibody (GE Healthcare Life Sciences, catalog 27-9421-01) for 1 hour at RT. After incubation with a rabbit anti–mouse IgG Cy3-conjugated secondary antibody (Jackson ImmunoResearch, catalog 315-165-003), cells were washed with PBS and refixed in PBS containing 4% PFA. Internalized phage particles were visualized with a standard fluorescence microscope (Nikon Ti-E Inverted Microscope).
NGS and data analysis. Sample preparation for NGS was performed as described (27). Briefly, plasmid DNA recovered from the selection outputs was amplified with a specific set of primers designed for MiSeq paired-end sequencing of the scFv heavy chain variable (VH) domains (24). The amplicons were sequenced with the MiSeqV2 kit for 500 cycles. Sequencing results were analyzed with the aid of the AbMining toolbox software package with default settings for quality filtering (27, 58). Identified HCDR3s were clustered at Hamming distance 1 and analyzed further in MS Excel (27, 58).
ScFv-Fc production. Monoclonal scFv antibody genes were subcloned either into the pHygro vector for scFv-Fc expression in CHO-S cells (Thermo Fisher Scientific) or into the yeast expression vector pDNL9 for expression into YVH10 S. cerevisiae yeast cells, as indicated (3). Both vectors allow the production of the scFv as a human IgG Fc fusion protein (26). The pHygro_scFv-Fc constructs were introduced into CHO-S cells by lipid-based transfection (FreeStyle Max Reagent, Gibco). Transient expression of the scFv-Fc fusion protein in the supernatant was achieved within 5 days of culturing in serum-free FreeStyle CHO Expression Medium (Gibco). The pDNL9_scFv-Fc construct was introduced into YVH10 cells by heat-shock and lithium acetate–based transformation. After 2 days of growth in selective media (selective drop-out media [SD] – case amino acids [CAA] + tryptophan [Trp]), yeast cells were induced for 3 days in induction media (selective growth [SGT] + Trp). The scFv-Fc proteins were recovered from the culture supernatant and were purified by affinity purification on Protein-G agarose (Roche Diagnostics). ELISA with the corresponding target was performed to test binding specificity of individually purified scFv-Fc. All cloning steps were monitored and verified by DNA sequencing analysis.
In vivo scFv-Fc targeting. Tumor homing was assessed by i.v. administration of the purified scFv-Fc in solution into tumor-bearing mice. Two doses of a 2.5 μM solution (100 and 200 μl) of purified scFv-Fc were administered into the tail vein of anesthetized tumor-bearing mice. scFv-Fcs were allowed to circulate for 6 minutes prior to full-body cardiac perfusion with PBS. Tumor and control organs were carefully removed to avoid cross-contaminantion and processed for either IHC or IF staining. IF was performed on frozen tissue sections using a Cy3-conjugated goat anti–human Fcγ IgG (Jackson ImmunoResearch, catalog 109-165-008) and DAPI for nuclei staining. Images were acquired with a Nikon Ti-E Inverted fluorescence microscope and processed with Adobe Photoshop.
In vitro cytotoxicity assays. Cell killing activity was measured in real time with the Xcelligence system (ACEA Biosciences). Freshly split tumor cells (25,000 cells/well) were cultured overnight (ON) in a 96-well electronic microtiter plate (E-plate 96, ACEA Biosciences) in 100 μl of complete culture medium. After 24 hours, increasing concentrations of the purified primary monoclonal scFv-Fcs were added to each microwell, followed by addition of 20 nM of secondary ADC reagents (Moradec LLC) linked to MMAF (Fab-αHFc-CL-MMAF, catalog AH-202AF-50), DMDM (Fab-αHFc-CL-DMDM, catalog AH-202DD-50), DM1 (Fab-αHFc-NC-DM1, catalog AH-203D1-50), or AAMT (Fab-αHFc-NC-AAMT, catalog AH-205AM-50), all obtained from Moradec. Cell index was measured every 30 minutes for 96 hours. Controls included primary scFv-Fc alone, drug-conjugated secondary antibody alone, and nontreated cells as indicated.
Statistics. Graphpad Prism software v.5.03 and Microsoft Excel were used to graph data as mean ± SD or SEM as indicated and to calculate P values by using homoscedastic (1-tailed) Student’s t tests. P < 0.05 was considered statistically significant.
Study approval. All experiments conformed to the University of New Mexico regulatory standards. The present studies in animals followed Animal Research Reporting: Reporting of In Vivo Experiments (ARRIVE) guidelines and were reviewed and approved by the IACUC of the University of New Mexico (Albuquerque, New Mexico, USA).
-
Author contributions
SD, FIS, FF, WA, ARMB, and RP designed experiments. SD, FIS, FF, DIS, GS, CAT, HN, and LAN performed experiments. SD, FIS, FF, WA, ARMB, and RP analyzed data. SD, FIS, FF, RLS, WA, ARMB, and RP wrote the initial manuscript, and all authors contributed edits.
-
Acknowledgments
This work was supported by grants from the National Cancer Institute (FIS, RP and WA), Department of Defense (RP and WA), NIH (ARMB), and the Gillson-Longenbaugh Foundation (RP and WA).
Address correspondence to: Wadih Arap or Renata Pasqualini, Rutgers Cancer Institute of New Jersey at University Hospital, Rutgers New Jersey Medical School, 205 South Orange Avenue, Newark, New Jersey, 07103, USA. Phone: 793.792.0366; Email: wadih.arap@rutgers.edu (W. Arap). Phone: 793.792.9958; Email: renata.pasqualini@rutgers.edu (R. Pasqualini). Or to: Andrew R.M. Bradbury, Specifica Inc., 1512 Pacheco Street, Suite A203, Santa Fe, New Mexico 87505, USA. Phone: 505.431.0065; Email: abradbury@specifica.bio.
-
Footnotes
Conflict of interest: SD, FIS, FF, WA, ARMB, and RP are inventors on patents (International Patent Application no. PCT/US2017/052661) on the technology and intellectual property reported here and, if licensing or commercialization occurs, they will be entitled to royalties.
Reference information: JCI Insight. 2018;3(9): e98305. https://doi.org/10.1172/jci.insight.98305.
-
References
- Ozawa MG, et al. Beyond receptor expression levels: the relevance of target accessibility in ligand-directed pharmacodelivery systems. Trends Cardiovasc Med. 2008;18(4):126–132.View this article via: CrossRef Google Scholar
- Carter P, Smith L, Ryan M. Identification and validation of cell surface antigens for antibody targeting in oncology. Endocr Relat Cancer. 2004;11(4):659–687.View this article via: CrossRef Google Scholar
- Ferrara F, et al. Using phage and yeast display to select hundreds of monoclonal antibodies: application to antigen 85, a tuberculosis biomarker. PLoS One. 2012;7(11):e49535.
- Arap W, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8(2):121–127.View this article via: CrossRef Google Scholar
- Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279(5349):377–380.
- Driessen WH, et al. On the synergistic effects of ligand-mediated and phage-intrinsic properties during in vivo selection. Adv Genet. 2010;69:115–133.View this article via: PubMed Google Scholar
- Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004;10(6):625–632.
- Kolonin MG, et al. Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB J. 2006;20(7):979–981.
- Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380(6572):364–366.
- Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest. 1998;102(2):430–437.
- Staquicini FI, et al. Vascular ligand-receptor mapping by direct combinatorial selection in cancer patients. Proc Natl Acad Sci USA. 2011;108(46):18637–18642.
- Staquicini FI, Moeller BJ, Arap W, Pasqualini R. Combinatorial vascular targeting in translational medicine. Proteomics Clin Appl. 2010;4(6-7):626–632.
- Staquicini FI, et al. Systemic combinatorial peptide selection yields a non-canonical iron-mimicry mechanism for targeting tumors in a mouse model of human glioblastoma. J Clin Invest. 2011;121(1):161–173.
- Krag DN, et al. Selection of tumor-binding ligands in cancer patients with phage display libraries. Cancer Res. 2006;66(15):7724–7733.
- Shukla GS, et al. Intravenous infusion of phage-displayed antibody library in human cancer patients: enrichment and cancer-specificity of tumor-homing phage-antibodies. Cancer Immunol Immunother. 2013;62(8):1397–1410.
- Deramchia K, et al. In vivo phage display to identify new human antibody fragments homing to atherosclerotic endothelial and subendothelial tissues [corrected]. Am J Pathol. 2012;180(6):2576–2589.
- Sánchez-Martín D, et al. Selection strategies for anticancer antibody discovery: searching off the beaten path. Trends Biotechnol. 2015;33(5):292–301.
- Staquicini FI, et al. Receptor tyrosine kinase EphA5 is a functional molecular target in human lung cancer. J Biol Chem. 2015;290(12):7345–7359.
- Arap MA, et al. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell. 2004;6(3):275–284.
- Dobroff AS, et al. Towards a transcriptome-based theranostic platform for unfavorable breast cancer phenotypes. Proc Natl Acad Sci USA. 2016;113(45):12780–12785.
- Ferrara F, et al. Targeted molecular-genetic imaging and ligand-directed therapy in aggressive variant prostate cancer. Proc Natl Acad Sci USA. 2016;113(45):12786–12791.View this article via: CrossRef Google Scholar
- Rauschert N, Brändlein S, Holzinger E, Hensel F, Müller-Hermelink HK, Vollmers HP. A new tumor-specific variant of GRP78 as target for antibody-based therapy. Lab Invest. 2008;88(4):375–386.
- Liu R, et al. Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin Cancer Res. 2013;19(24):6802–6811.
- de Ridder GG, Ray R, Pizzo SV. A murine monoclonal antibody directed against the carboxyl-terminal domain of GRP78 suppresses melanoma growth in mice. Melanoma Res. 2012;22(3):225–235.
- Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol. 2000;18(1):75–80.
- Ferrara F, et al. Recombinant renewable polyclonal antibodies. MAbs. 2015;7(1):32–41.
- D’Angelo S, et al. The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. MAbs. 2014;6(1):160–172.
- Barnhart KF, et al. A peptidomimetic targeting white fat causes weight loss and improved insulin resistance in obese monkeys. Sci Transl Med. 2011;3(108):108ra112. View this article via: PubMed Google Scholar
- Christianson DR, Ozawa MG, Pasqualini R, Arap W. Techniques to decipher molecular diversity by phage display. Methods Mol Biol. 2007;357:385–406.View this article via: PubMed Google Scholar
- Dias-Neto E, et al. Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS One. 2009;4(12):e8338.
- Hajitou A, Pasqualini R, Arap W. Vascular targeting: recent advances and therapeutic perspectives. Trends Cardiovasc Med. 2006;16(3):80–88.
- Kolonin M, Pasqualini R, Arap W. Molecular addresses in blood vessels as targets for therapy. Curr Opin Chem Biol. 2001;5(3):308–313.
- Pasqualini R, Arap W. Profiling the molecular diversity of blood vessels. Cold Spring Harb Symp Quant Biol. 2002;67:223–225.
- Chasteen L, Ayriss J, Pavlik P, Bradbury AR. Eliminating helper phage from phage display. Nucleic Acids Res. 2006;34(21):e145.
- Phipps ML, et al. Beyond Helper Phage: Using “Helper Cells” to Select Peptide Affinity Ligands. PLoS One. 2016;11(9):e0160940.
- Gabbard J, et al. A humanized anti-M2 scFv shows protective in vitro activity against influenza. Protein Eng Des Sel. 2009;22(3):189–198.View this article via: PubMed Google Scholar
- Di Niro R, et al. Construction of miniantibodies for the in vivo study of human autoimmune diseases in animal models. BMC Biotechnol. 2007;7:46. View this article via: PubMed Google Scholar
- Llewellyn DH, Roderick HL, Rose S. KDEL receptor expression is not coordinatedly up-regulated with ER stress-induced reticuloplasmin expression in HeLa cells. Biochem Biophys Res Commun. 1997;240(1):36–40.
- Ni M, Zhang Y, Lee AS. Beyond the endoplasmic reticulum: atypical GRP78 in cell viability, signalling and therapeutic targeting. Biochem J. 2011;434(2):181–188.
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–497.
- Brüggemann M, et al. Human antibody production in transgenic animals. Arch Immunol Ther Exp (Warsz). 2015;63(2):101–108.
- Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321(6069):522–525.
- Morrison SL, Johnson MJ, Herzenberg LA, Oi VT. Chimeric human antibody molecules: mouse antigen-binding domains with human constant region domains. Proc Natl Acad Sci USA. 1984;81(21):6851–6855.
- Queen C, et al. A humanized antibody that binds to the interleukin 2 receptor. Proc Natl Acad Sci USA. 1989;86(24):10029–10033.
- Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature. 1988;332(6162):323–327.
- Gerhard W, Croce CM, Lopes D, Koprowski H. Repertoire of antiviral antibodies expressed by somatic cell hybrids. Proc Natl Acad Sci USA. 1978;75(3):1510–1514.
- Koprowski H, Gerhard W, Croce CM. Production of antibodies against influenza virus by somatic cell hybrids between mouse myeloma and primed spleen cells. Proc Natl Acad Sci USA. 1977;74(7):2985–2988.
- Pasqualini R, Arap W. Hybridoma-free generation of monoclonal antibodies. Proc Natl Acad Sci USA. 2004;101(1):257–259.
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222(3):581–597.
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15(6):553–557.
- Fu Y, Li J, Lee AS. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen starvation-induced apoptosis. Cancer Res. 2007;67(8):3734–3740.
- Miao YR, et al. Inhibition of established micrometastases by targeted drug delivery via cell surface-associated GRP78. Clin Cancer Res. 2013;19(8):2107–2116.
- Quinones QJ, de Ridder GG, Pizzo SV. GRP78: a chaperone with diverse roles beyond the endoplasmic reticulum. Histol Histopathol. 2008;23(11):1409–1416.View this article via: PubMed Google Scholar
- Sato M, Yao VJ, Arap W, Pasqualini R. GRP78 signaling hub a receptor for targeted tumor therapy. Adv Genet. 2010;69:97–114.View this article via: PubMed Google Scholar
- Hajitou A, et al. FGF-3 and FGF-4 elicit distinct oncogenic properties in mouse mammary myoepithelial cells. Oncogene. 1998;17(16):2059–2071.
- Feldhaus MJ, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21(2):163–170.
- Boder ET, Wittrup KD. Optimal screening of surface-displayed polypeptide libraries. Biotechnol Prog. 1998;14(1):55–62.
- D’Angelo S, Kumar S, Naranjo L, Ferrara F, Kiss C, Bradbury AR. From deep sequencing to actual clones. Protein Eng Des Sel. 2014;27(10):301–307.
- Ozawa MG, et al. Beyond receptor expression levels: the relevance of target accessibility in ligand-directed pharmacodelivery systems. Trends Cardiovasc Med. 2008;18(4):126–132.
-
Version history
- Version 1 (May 3, 2018): Electronic publication








