Research ArticleCell biologyDermatologyMetabolism
Open Access |  10.1172/jci.insight.194188
10.1172/jci.insight.194188
A multiomics analysis identifies retinol metabolism in fibroblasts as a key pathway in wound healing
Till Wüstemann,1 Elizabeta Madzharova,2 Mateusz S. Wietecha,1,3 Norbert B. Ghyselinck,4 Marcus Höring,5 Gerhard Liebisch,5 Nicola Zamboni,6 Ulrich auf dem Keller,2 and Sabine Werner1
1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by Wüstemann, T. in: PubMed | Google Scholar
1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by Madzharova, E. in: PubMed | Google Scholar
1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by Wietecha, M. in: PubMed | Google Scholar
1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by Ghyselinck, N. in: PubMed | Google Scholar
1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by
Höring, M.
in:
PubMed
|
Google Scholar
|

1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by
Liebisch, G.
in:
PubMed
|
Google Scholar
|

1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by Zamboni, N. in: PubMed | Google Scholar
1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by auf dem Keller, U. in: PubMed | Google Scholar
1Institute of Molecular Health Sciences, Department of Biology, ETH Zurich, Zurich, Switzerland.
2Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.
3Department of Oral Biology, College of Dentistry, University of Illinois Chicago, Chicago, Illinois, USA.
4Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Département de Génétique Fonctionnelle et Cancer, Centre National de la Recherche Scientifique (CNRS UMR7104), Institut National de la Santé et de la Recherche Médicale (INSERM U1258), Université de Strasbourg (UNISTRA), Illkirch Cedex, France.
5Institute of Clinical Chemistry and Laboratory Medicine, University of Regensburg, Regensburg, Germany.
6Institute of Molecular Systems Biology, Department of Biology, ETH Zurich, Zurich, Switzerland.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
Authorship note: UADK is deceased.
Find articles by Werner, S. in: PubMed | Google Scholar
Authorship note: UADK is deceased.
Published October 16, 2025 - More info
JCI Insight. 2025;10(22):e194188. https://doi.org/10.1172/jci.insight.194188.
© 2025 Wüstemann et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: April 4, 2025; Accepted: October 7, 2025
-
Abstract
Impaired wound healing poses a major and increasingly frequent health problem. Among the key players in the healing process are fibroblasts, but their metabolic profile in healing wounds is largely unknown. Using a combination of transcriptomics, targeted proteomics, and metabolomics, we identified retinol metabolism as a top regulated pathway in wound fibroblasts. This is functionally relevant, since even a mild retinol deficiency caused a delay in wound closure and reepithelialization, which mainly resulted from misdirected keratinocyte migration on the new granulation tissue. Quantitative proteomics identified integrin subunit α11 as a less abundant protein in wounds of mice subjected to a retinol-deficient diet. Reduced levels of this fibroblast-specific protein likely altered the granulation tissue matrix, which in turn affected reepithelialization. These results provide a comprehensive overview of the transcriptome, proteome, and metabolome of wound fibroblasts and identify retinol metabolism in fibroblasts as a key regulator of tissue repair.
-
Introduction
Wound healing requires the coordinated action of different cell types to achieve the restoration of tissue integrity (1). It involves 3 partially overlapping phases, including blood clotting and inflammation, new tissue formation, and tissue remodeling (2). One of the key cell types involved in skin wound healing are fibroblasts. During the proliferative phase of wound healing, they migrate into the wound area, proliferate, and deposit the majority of extracellular matrix (ECM) proteins. This allows keratinocytes at the wound edge to migrate on top of the newly formed granulation tissue and to seal the wound with a new epidermis, which reestablishes the important skin barrier. While recent studies have focused on cell fate, expression profiles, and spatial distribution of fibroblasts in skin wounds (3–9), much less is known regarding their metabolic features. A drastically reshaped environment, which wounds pose, heavily alters cellular metabolism and challenges the cells in the wound tissue to optimally allocate resources and metabolites (10, 11). Previous studies identified the necessity of functional bioenergetic pathways for successful wound healing and highlighted the importance of glycolysis in macrophages for efficient healing (12, 13). In addition, distinct metabolic profiles were detected for wound tissue compared with normal skin (14). In fibrotic skin, metabolic changes in fibroblasts altered their ECM deposition and arrangement (15). However, the metabolome of fibroblasts during wound healing has not been analyzed in an unbiased way, though these cells regulate their surrounding ECM depending on their metabolic state (15).
To fill this knowledge gap, we characterized the fibroblast metabolome of normal mouse skin and of wounds during the phase of granulation tissue formation and compared it with their transcriptome. Together with targeted and quantitative proteomics data of normal and wounded skin and functional wound healing studies, these data identify retinol metabolism as a key pathway in wound healing.
-
Results
Wounding activates the retinol pathway in fibroblasts. To identify changes in the fibroblast metabolome and transcriptome that occur upon wounding, we FACS-sorted fibroblasts from nonwounded skin and 5-day full-thickness excisional wounds of wild-type mice as described before (16) using antibodies against the pan-fibroblast marker platelet-derived growth factor receptor α (Pdgfrα; CD140a), which is not expressed by dermal endothelial cells (4, 17). The 5-day time point was chosen, because it represents the peak of the phase of new tissue formation (18). We gated for live CD45–F4/80–CD11b– nonimmune cells to exclude immune cells that occasionally express Pdgfra (19) and then for Pdgfrα+ cells. Cell lysates were subjected to untargeted metabolomics analysis (Figure 1A). Skin and wound tissue were treated identically to exclude alterations in metabolites that result from the tissue dissociation and sorting procedure (20). We also reanalyzed our previously published RNA-Seq data from skin and 5-day wound fibroblasts of mice of the same genetic background for genes that are regulated by wounding in wild-type mice (Figure 1A) (16).
 Figure 1
Figure 1Retinol metabolism is strongly regulated by skin wounding. (A) Outline of the experimental setup. Pdgfrα+ fibroblasts were sorted from normal skin and 5-day wounds and subjected to RNA-Seq and metabolomics analysis. N = 3 mice per experiment. Created in BioRender. Werner, S. (2025) https://BioRender.com/aaw9vrp (B) Volcano plot showing differentially abundant metabolites in wound vs. normal skin fibroblasts. Colored points indicate metabolites with significantly differential abundance (P ≤ 0.05); diamond-shaped points indicate metabolites that were confirmed in an independent experiment. N = 3 mice per experiment. (C) Volcano plot showing differentially expressed genes in wound vs. normal skin fibroblasts based on bulk RNA-Seq analysis of FACS-sorted fibroblasts (16). Colored points indicate significantly differentially expressed genes (Bonferroni-corrected P ≤ 0.05, |log2(FC)| ≥ 2). N = 3 mice. Rdh10, retinol dehydrogenase 10; Rbp4, retinol-binding protein 4; Stra6, stimulated by retinoic acid 6. (D) Integrated pathway analysis of RNA-Seq and metabolomics data sets using MetaboAnalyst 6.0. Colored points indicate significantly altered pathways (P ≤ 0.05). (E) Schematic overview of retinol metabolism with highly relevant metabolites and genes. Metabolites with a strikethrough pattern were not detected in the metabolomics experiment. Members of the respective enzyme families that were significantly differentially expressed based on RNA-Seq data are featured (uncorrected P ≤ 0.05). Size of the circles corresponds to the log10(RPKM+0.95) (0.95 was added to all values to avoid negative results of the logarithm), and color represents the log2(fold change) of the genes (red: increased expression in wound vs. normal skin fibroblasts; blue: reduced expression in wound vs. normal skin fibroblasts). RPKM, reads per kilobase million.
Twenty-one unique metabolites were differentially abundant between normal and wounded skin and detected in 2 independent wound healing experiments (Figure 1B). RNA-Seq data confirmed the efficient separation of fibroblasts from endothelial cells as revealed by the low levels of the endothelial-specific Pecam1, Kdr, Cdh5, Cldn5, and Vwf transcripts (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.194188DS1). There were strong alterations in the fibroblast transcriptome upon wounding (Figure 1C). Gene ontology overrepresentation analysis of the RNA-Seq data identified multiple pathways involved in ECM constitution and binding that were significantly overrepresented in wound fibroblasts (Supplemental Figure 1B).
The identical preparation of fibroblasts for the metabolomics (this study) and transcriptomic analyses (16) allowed us to integrate these data sets and to determine how the expression of metabolism-associated genes matches with the metabolites present in the cell. There was a strong correlation between the wound-enriched metabolites and mRNAs encoding the enzymes required for their production or proteins involved in their transport. We focused on the significance of the alteration and the pathway impact, which captures the relevance of a pathway. Among the significantly altered pathways, retinol (vitamin A) metabolism scored highest on pathway impact (Figure 1D).
Analysis of retinol metabolism genes and metabolites within the acquired data revealed strong alterations in multiple components of this pathway (Figure 1E and Supplemental Table 1). Genes that are more highly expressed in wound versus normal skin fibroblasts encode proteins involved in storage and trafficking of retinoids, such as retinol-binding protein 4 (Rbp4), which is required for the release from its stores in the liver and transport in the serum, the retinol receptor and transporter stimulated by retinoic acid 6 (Stra6), which promotes uptake of retinol by cells (21), and the intracellular Rbp1, which is responsible for retinol transport to acyltransferases for storage as retinol esters or to dehydrogenases for stepwise oxidization to retinoic acid (RA) (22–24). Similarly, Crabp1, which encodes the cellular RA-binding protein 1, was expressed at higher levels in wound fibroblasts. This intracellular RA chaperone is responsible for RA transfer into the nucleus to induce gene expression changes via binding to RA receptor (RAR)/retinoid X receptor heterodimers (Figure 1E).
The most strongly regulated step in the retinol pathway was the oxidation of retinol. Several genes of this rate-defining step of retinol metabolism (25) were strongly regulated upon wounding, e.g., retinol dehydrogenases 8 and 10 (Rdh8 and Rdh10; upregulated) and short-chain dehydrogenase/reductase family 16C members 5 and 6 (Sdr16c5 and Sdr16c6; downregulated). Expression of most of the enzymes responsible for the oxidation of retinal to RA was downregulated, in particular aldehyde dehydrogenase 1 family member A1 (Aldh1a1). These findings suggest enhanced uptake of retinol by wound fibroblasts, its accumulation in these cells, and a concomitant reduction in the expression of genes associated with retinol metabolization to RA. This reduction may be a compensatory mechanism to maintain appropriate levels of RA, which activates RAR-mediated gene expression.
Together, these results identify retinol metabolism as a key pathway that is activated in wound fibroblasts. Analysis of published single-cell RNA-Seq (scRNA-Seq) data from mouse excisional wounds at different stages (26) confirmed this finding and also revealed that this pathway is mainly activated between day 3 and 7 after wounding, especially in fibroblasts (Supplemental Figure 1, C and D).
Retinol metabolism genes are activated by retinoids in cultured fibroblasts. We next searched for potential regulators of retinol metabolism in the wound environment. Because wound fibroblasts are exposed to serum, which induces a wound fibroblast phenotype (27), and because serum contains high levels of retinol (in mice and humans around 1 μmol/L) (28, 29), we tested the effect of serum and of retinol itself on the expression of retinol metabolism genes. We focused on STRA6/Stra6, RBP1/Rbp1, RPB4/Rbp4, and CRABP1/Crabp1, whose expression correlated with the increase in retinol metabolism in wound fibroblasts, as well as on the gene encoding RA receptor β (RARB/Rarb), a main component and direct target of RAR signaling (30, 31). To separate potential effects of peptide growth factors from retinol and other lipophilic components in the serum, we cultured human primary fibroblasts in medium with either nontreated FBS or FBS that had been preincubated with dextran-coated, activated charcoal (referred to as charcoal-stripped). Activated charcoal adsorbs lipophilic molecules, including retinoids. Although nontreated FBS induced a strong expression of retinol metabolism–associated genes after 24 hours, the effect on RARB and STRA6 was significantly lower with charcoal-stripped FBS (Figure 2A). The reduction was similar to the effect seen with the pan-RAR inhibitor AGN194310 and with the pan-RAR inverse agonist AGN193109, which counteracts the activated and baseline expression of retinol metabolism genes in a dose-dependent manner (Figure 2A and Supplemental Figure 2A) (32). The reduction by charcoal-stripped FBS was almost fully rescued by retinol supplementation of the medium (Figure 2A), indicating that the effect of FBS on the expression of retinol metabolism–associated genes is largely mediated by retinol. Retinol treatment of primary mouse fibroblasts or primary human skin fibroblasts from 2 donors indeed promoted the expression of multiple retinol metabolism genes (Figure 2, B and C). The most responsive gene was RARB, the only RAR gene that was directly activated by retinoids (Supplemental Figure 2B). With the exception of RDH10, the regulation by retinoids in vitro was similar to the regulation seen in wound fibroblasts. TNF-α, which is highly expressed in early murine skin wounds (33), had only a minor or no effect on the expression of the different retinol metabolism genes (Supplemental Figure 2C). This was also the case for menadione (Supplemental Figure 2D), which induces the production of ROS. In skin wounds, ROS are mainly produced by immune cells during their oxidative burst.
 Figure 2
Figure 2Retinol metabolism genes are regulated by retinoids in cultured fibroblasts. (A) Real-time quantitative PCR (RT-qPCR) for STRA6 and RARB relative to RPL27 using RNA from primary human fibroblasts, treated with 1 μM retinol (ROL), 0.1 μM pan-RAR inverse agonist (AGN193109), 0.1 μM pan-RAR inhibitor (AGN194310), charcoal-stripped FBS (Stripped FBS), or untreated FBS (for samples without Stripped FBS treatment) for 24 hours. N = 6 cultures from 1 donor per treatment group. (B) RT-qPCR for Stra6, Rbp1, Lrat, and Rdh10 relative to Rps29 using RNA from primary mouse fibroblasts, treated with 1 μM ROL for 24 hours after 48 hours of starvation in medium supplemented with 1% FBS. N = 5–6 cultures from 2–3 mice per treatment group. (C) RT-qPCR for STRA6 and RBP1 using RNA from primary human fibroblasts from 2 different donors (1 and 2), treated with 1 μM ROL, retinal (RAL), or RA for 24 hours after 48 hours of starvation in serum-free medium. N = 3 cultures per donor and treatment group. Graphs show mean ± SD. **P < 0.01, ****P < 0.0001, 1-way ANOVA, Šídák’s multiple comparisons test (A and C), or Mann-Whitney test (B).
Taken together, these results suggest that the increased expression of retinol metabolism genes in wound fibroblasts results at least in part from their exposure to serum. They also identify retinol itself as a major serum component that mediates this effect.
Loss of Stra6 has no major effect on wound healing in mice. In tissues that require high concentrations of retinol, such as the retina, STRA6 facilitates the transport of retinol across the plasma membrane (35). The increased expression of Stra6 in wound fibroblasts suggested that retinol uptake via diffusion may not be sufficient and that fibroblasts may require additional retinol uptake via Stra6 during wound healing. To test this hypothesis, we subjected mice with a global knockout of Stra6 (Stra6-KO mice) to full-thickness excisional wounding (Figure 3, A and B). These mice have a vision defect but do not exhibit additional abnormalities under nonchallenged conditions (Supplemental Figure 3A and ref. 35). Histomorphometric analysis (Figure 3C) of H&E-stained sections from the middle of the wounds did not reveal significant alterations in reepithelialization, wound contraction, or neo-epidermal thickness (Figure 3D). Analysis of the hydroxyproline content of wound lysates showed a mild, but nonsignificant, reduction of the collagen levels in the knockout mice. Consistently, deposition of collagen in the granulation tissue and maturation of young to mature collagen, as examined by Herovici staining of wound sections, was unaltered (Figure 3E). Flow cytometry analysis of Pdgfrα+ fibroblasts further showed that the number of live wound fibroblasts was not affected, and primary skin fibroblasts isolated from Stra6-KO mice had a similar proliferation rate as fibroblasts from wild-type mice within 3 days of culture (Supplemental Figure 3, B and C). RT-qPCR analysis of RNA from whole wounds for Inhba, which encodes activin A, an important regulator of wound healing and matrix production (16); Col1a1, encoding the major skin collagen; and Socs3, one of the main targets of the Stra6/Jak2/Stat5 pathway that is activated by Stra6 (36), showed increased expression of all 3 genes in wounded versus normal skin but no differences between genotypes (Supplemental Figure 3D). Finally, the upregulation of retinol metabolism–associated genes in wound fibroblasts was not affected by the knockout, except for Stra6 itself (Supplemental Figure 3D). Together, these results reveal that loss of Stra6 does not result in major wound healing abnormalities in mice.
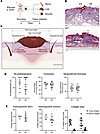 Figure 3
Figure 3Stra6 is dispensable for wound healing in mice. (A) Schematic outline of the experimental setup. Created in BioRender. Werner, S. (2025) https://BioRender.com/fxc8gmk (B) Representative images of H&E-stained 5-day wound sections from wild-type (WT) and Stra6–/– mice. G, granulation tissue; HE, hyperproliferative wound epidermis; Es, eschar. (C) Illustration of wound histomorphometry measurements. Created in BioRender. Werner, S. (2025) https://BioRender.com/714lbmt (D) Histomorphometric parameters based on H&E-stained wound sections. N = 11–13 mice per genotype (1 wound per mouse). (E) Collagen analysis by quantification of hydroxyproline in acid hydrolyzed skin and wound tissues and by quantification of Herovici- (graph “Collagen area”) stained wound sections. N = 4–13 mice per genotype (1 wound per mouse). Graphs show mean ± SD. Scale bars: 100 μm.
Mild vitamin A deficiency delays wound healing. To further reduce the availability of retinol for fibroblasts and other cells in the wound, we subjected wild-type mice to a vitamin A–deficient diet (VAD) for 6 weeks prior to wounding (Figure 4A) (37). The short diet had no adverse effect and did not lead to significant changes in body weight (Supplemental Figure 4, A and B). The levels of liver retinyl palmitate, one of the main storage forms of retinol, were mildly reduced, likely reflecting the onset of retinol deficiency. However, the difference was not statistically significant because of the high variability in the control mice (Supplemental Figure 4C). As expected from published studies (29, 38), serum retinol levels were not yet reduced at this stage (Supplemental Figure 4D).
 Figure 4
Figure 4Vitamin A deficiency delays wound healing in mice. (A) Schematic outline of the experimental setup. VAD, vitamin A–deficient diet. Created in BioRender. Werner, S. (2025) https://BioRender.com/1psvb1s (B) Representative images of H&E-stained sections from 5-day wounds of chow- and VAD-fed mice. G, granulation tissue; HE, hyperproliferative wound epidermis; Es, eschar. Black arrowheads mark wound tongues; chow wound is closed. (C and D) Histomorphometric parameters based on H&E-stained wound sections of mice fed with VAD or chow diet. N = 21–23 mice per treatment group (1 wound per mouse) from 3 independent experiments. Effective migration quantifies the directional keratinocyte migration that contributes to wound closure. (E) Percentage of Ki67-positive nuclei and Ki67-positive nuclei/mm2 tissue in the epidermis and granulation tissue of wounds from mice fed with VAD or chow diet. N = 8 mice per treatment group (1 wound per mouse). (F) Percentage of Ki67-positive nuclei and Ki67-positive nuclei per mm2 tissue in hair follicles at the wound edge of mice fed with VAD or chow diet. N = 8 mice per treatment group (1 wound per mouse). Graphs show mean ± SD. *P < 0.05, **P < 0.01 (Mann-Whitney test). Scale bars: 250 μm.
Histological and histomorphometric analysis of wound sections showed a significant reduction in reepithelialization and wound closure in VAD- versus chow-fed animals (Figure 4, B and C), while the length of the epithelial tongue, which reflects the extent of migration, was not significantly changed at this time point (Figure 4C). However, the directional migration toward the middle of the wound was significantly impaired in VAD-fed mice (“effective migration”; Figure 4C), providing an explanation for the delayed wound closure. This may point toward impaired migration on the matrix of the newly formed granulation tissue. As contraction strongly contributes to wound closure in mice (39), the normal wound contraction in VAD-treated mice (Figure 4D) allowed an efficient healing process in spite of the impaired reepithelialization. The granulation tissue area in 5-day wounds was similar in mice of both treatment groups (Figure 4D), though the VAD-fed mice showed a significant increase in adipose tissue in the wounds as revealed by semiquantitative blinded wound scoring by investigators (Figure 4D). Epidermal cell proliferation, which is reflected by the neo-epidermal thickness and the number of Ki67-positive epidermal cells (Figure 4, D and E), was not affected by VAD treatment. Furthermore, there were no differences in the rate of proliferation of cells in the granulation tissue and in the hair follicles at the wound edge (Figure 4, E and F), the collagen-positive wound area, the ratio between Picrosirius red–stained collagens and fast green FCF–stained noncollagenous peptides, the hydroxyproline content of the wounds, and the extent of neovascularization (shown by staining with the pan–endothelial cell marker plasmalemma vesicle associated protein, MECA-32) (Supplemental Figure 4, E–H). Despite the significant increase in adipocyte area in the granulation tissue, no significant difference in the content of 16 different lipids were detected (Supplemental Figure 5A).
Taking together, these results show that mild vitamin A deficiency induces defects in wound reepithelialization, which are at least in part mediated by misdirected migration of keratinocytes.
Wounding alters the abundance of retinol metabolism proteins in the skin. Although obvious alterations in the granulation tissue were not detected, more subtle changes cannot be excluded by these histological data. Therefore, we determined the effect of VAD treatment on the proteome of normal and wounded skin. We first performed targeted proteomics using whole skin and 5-day wound lysates to determine if the increased expression of retinol metabolism genes in wounds translates into alterations at the protein level (Figure 5A). We designed 65 peptides corresponding to 22 proteins associated with retinol metabolism. Thirty-six of these peptides were detected in the skin/wound analysis, of which 34 mapped to a total of 14 proteins. Among the proteins for which 2 peptides were detected, 5 showed a significantly differential abundance in wound versus normal skin tissue (Figure 5B). Consistent with the RNA-Seq data from isolated fibroblasts, Rbp1, Rbp4, and Rabp1 (encoded by the Crabp1 gene) were more abundant in wound tissue. The abundance of Aldh1a1, one of the rate-limiting proteins in the oxidation of retinaldehyde to RA, was significantly lower in wounds, again following the expression changes observed by RNA-Seq. Awat1, which is involved in the storage of retinyl esters, was significantly more abundant in wounds, while its mRNA was downregulated (Figure 5B). This finding indicates regulation at the protein level or a disparate regulation in fibroblasts compared with other wound cells that were also analyzed in the proteomics experiment. VAD did not significantly alter the abundance of the proteins that we identified by targeted proteomics. Together, these results confirm the RNA-Seq and metabolomics data but do not point to a regulation of retinol metabolism proteins by VAD.
 Figure 5
Figure 5Wounding alters the abundance of retinol metabolism proteins in the skin, and VAD alters the wound proteome. (A) Outline of the experimental setup. Created in BioRender. Werner, S. (2025) https://BioRender.com/z88rn2k (B) Quantification of significantly differentially abundant retinol metabolism proteins in skin and wound samples of chow-fed or VAD-treated mice based on targeted proteomics analysis (SureQuant). N = 5-6 mice. Awat1, acyl-CoA wax alcohol acyltransferase 1. (C) Volcano plot showing differentially abundant proteins in lysates from 5-day wounds of chow-fed or VAD-treated mice based on untargeted DIA proteomics analysis. Colored points indicate significantly differentially abundant proteins (P < 0.05, |logFC| > 0.15). N = 5 mice. (D) Quantification of top hits based on DIA analysis in C. N = 5 mice. (E) Western blot for α-SMA, periostin, and vinculin (loading control) using total wound lysates of chow-fed or VAD-treated mice. (F) Quantification of the band intensities of E and representation as ratio to the respective vinculin loading control. N = 5 mice per treatment group. (G) Representative images of immunofluorescence stainings for α-SMA (Cy3, orange) of 5-day wound sections of chow-fed and VAD-treated mice from an independent experiment. Hoechst was used to stain nuclei (blue). The area indicated with a white rectangle is shown at higher magnification at the right-hand side. Scale bars: 200 μm (overview) or 20 μm (detail). (H) Percentage of the α-SMA+ area in the granulation tissue of 5-day wound sections (G), determined by quantification of the Cy3-positive area and subtracting the background from the secondary antibody control. N = 5–6 mice per treatment group. Graphs show mean ± SD. *P < 0.05, **P < 0.01, ****P < 0.0001. Two-way ANOVA, Šídák’s multiple comparisons test (B), or Mann-Whitney test (D, F, and H).
Vitamin A deficiency alters the granulation tissue proteome. To determine if VAD affects the wound proteome, we performed quantitative, untargeted data-independent acquisition (DIA) proteomics. A total of 44,197 peptides were identified, which related to 4,451 identified proteins, including some with a significantly differential abundance in wounds of VAD- versus chow-fed controls (P ≤ 0.05, |logFC| ≥ 0.15) (Figure 5C). The top hits were Zhx3, a transcriptional repressor with a documented function in the regulation of senescence (40); Itga11, a protein important for adhesion of cells, including myofibroblasts, to the ECM and for wound strength (41); and Ggcx, which catalyzes posttranslational modifications of vitamin K–dependent proteins and is involved in the regulation of glucose metabolism in osteoblasts (42) (Figure 5D). Consistently, Western blot analysis revealed a mild reduction in the levels of the myofibroblast marker α–smooth muscle actin (α-SMA) and of periostin, a matricellular protein that is highly expressed in wounds and fibrotic scars and a potential target for γ-carboxylation by Ggcx (Figure 5, E and F) (43). Immunostaining of wound sections from an independent wounding experiment confirmed that the α-SMA+ area in the granulation tissue was significantly reduced in VAD-treated mice (Figure 5, G and H). Together, these findings support the hypothesis that VAD alters the composition of the wound granulation tissue.
We next analyzed published scRNA-Seq data, which had been obtained at different time points post wounding and with tissue of different distance from the middle of the wound (26), to determine if the identified hits are expressed by wound fibroblasts. Figure 6A depicts a uniform manifold approximation and projection (UMAP) visualization of the different cell types, subtypes, and spatiotemporal labels defined in the scRNA-Seq reanalysis. Zhx3, Itga11, and Ggcx transcripts were mainly detected in fibroblasts (Figure 6B). While Itga11 was exclusively expressed in cells with an early fibroblast expression profile, Ggcx was expressed in early, late, and activated fibroblasts (Figure 6C). Zhx3 mRNA was highly enriched in cells with a late fibroblast profile but also showed moderate expression levels in endothelial cells. We re-created spatiotemporal expression profiles of the 3 proteomics hits in all cells and in fibroblasts. Expression of Zhx3 increased on day 7 in the outer areas of the wound and was strongly expressed until day 14. Itga11 expression peaked near the wound center on day 7 after wounding. Ggcx showed the highest expression in the outer areas of the wound (Figure 6, D and E).
 Figure 6
Figure 6Proteomics hit proteins are mainly expressed in wound fibroblasts. (A) Uniform manifold approximation and projection (UMAP) visualization of wound cells and their annotations to cell types (left panel), subtypes (middle panel), and spatiotemporal labels (right panel) based on a mouse wound dataset (26). (B) Breakdown of expression of Zhx3, Itga11, and Ggcx in all annotated cell types. Dot size correlates with the percentage of cells that express the gene of interest, while color represents their average expression levels. (C) Breakdown of expression of Zhx3, Itga11, and Ggcx in all annotated cell subtypes (left) and specifically in the annotated fibroblast subtypes (right). Dot size correlates with the percentage of cells that express the gene of interest, while color represents their average expression levels. (D and E) Spatiotemporal expression of Zhx3, Ggcx, and Itga11 in all cells (D) and in the fibroblast subcluster (E). Space on the y axis refers to increasingly large rings around the wound center; wounds were sampled on day 1, 3, 7, and 14 as depicted on the x axis. The number in the square indicates the percentage of the cell subpopulation among all cells. Background color indicates relative change compared with the unwounded state (red: increase, blue: decrease, white: no change).
Finally, we tested whether the 3 hit proteins are direct targets of RA signaling. Treatment of primary human fibroblasts or immortalized mouse fibroblasts with retinol, RA, or the pan-RAR inverse agonist AGN193109 in the presence or absence of serum did not affect the expression of GGCX and ZHX3, while RARB and CRABP1 were strongly regulated. ITGA11 expression was even mildly reduced by RAR signaling in primary human fibroblasts (Supplemental Figure 6, A–C). The 3 hit genes were also not consistently regulated by retinols in cultured mouse fibroblasts (Supplemental Figure 6, D and E). Together, these findings indicate that the altered abundance of GGCX, ITGA11, and ZHX3 in wounds of VAD-treated mice results from alterations in the fibroblast phenotype and not from a direct effect of retinoids on their expression. The delayed reepithelialization of wounds in VAD-treated mice is a likely consequence of the alterations in fibroblast-specific proteins and concomitant changes in the granulation tissue composition.
-
Discussion
In this study, we identified retinol metabolism as a key pathway in wound healing. Notably, we observed enrichment of retinol and of retinol and RA binding proteins. The genes for 2 of them, Crabp1 and Fabp5, had been identified as markers for regenerative fibroblasts in scRNA-Seq analyses of mouse wounds, suggesting that the increase in retinol metabolism is important for repair (9, 44, 45). Analysis of published RNA-Seq data from mouse wounds at different time points confirmed the activation of retinol metabolism–associated genes in the proliferative phase of wound healing, in particular in fibroblasts. Consistent with an important role of retinol metabolism in wound healing, the efficiently regenerating MRL/MpJ mouse line exhibits increased expression of genes involved in retinol metabolism (46). Vice versa, serum retinol levels were reduced in patients with burn wounds or chronic leg ulcers (47, 48). Therefore, retinoids have been tested for their efficacy to stimulate wound healing (49). In most studies, topical retinoid treatment promoted repair in healing-impaired male or female mice or rats (50–52). In wounds of healthy pigs, topical tretinoin (all-trans RA) treatment retarded while tretinoin pretreatment promoted healing (53). In a human study, short-contact treatment of chronic leg ulcers with tretinoin promoted granulation tissue formation in male and female patients (54). Different effects of retinol and its derivatives on myofibroblast differentiation were also observed. While VAD reduced the number of myofibroblasts in mouse wounds in vivo (this study), retinoids inhibited and RAR antagonists promoted contraction of mouse fascia explant cultures (55). In a bleomycin-induced skin fibrosis model, inhibition of RA catabolism and the resulting increase in RA signaling inhibited fibrosis and shifted fibroblast populations towards a regenerative state (56). Therefore, different fibroblast subtypes may respond differently to retinoids, which could be further influenced by the in vivo environment. Together, these results indicate that retinol metabolism and RA signaling in wounds must be spatially and temporally tightly regulated and that they depend on the underlying pathology.
In contrast to these insightful treatment studies, the consequences of retinol deficiency for wound healing are still largely unknown. Here we show that the knockout of Stra6 has no major effect on wound healing in mice, suggesting that retinol uptake by diffusion is largely sufficient. While Stra6-KO mice only show a disruption of the visual cycle (35), truncating STRA6 mutations in humans are associated with severe organ malformations and growth retardations (57, 58). These findings point to a different importance of Stra6/STRA6 in mouse versus human tissue and suggest that STRA6 deficiency may have more severe consequences for wound healing in humans.
To further decrease the availability of retinol for wound fibroblasts, we subjected wild-type mice to a 6-week VAD. We refrained from prolonging the diet and thereby inducing a pathological vitamin A deficiency, as the resulting systemic abnormalities and malady would lead to unspecific wound healing defects. Serum retinol levels only decline after the liver stores of retinyl esters are depleted, but consequences of vitamin A deficiency were already observed before significant changes in total serum retinol were detected (29, 38). This may be explained by differences in the uptake of different retinol complexes. While retinol is almost exclusively bound to Rbp4 under fasting conditions, it is found in the form of retinyl esters in chylomicrons postprandially. Most of these chylomicrons are taken up by the liver, where retinol is stored for later distribution through Rbp4. However, some extrahepatic tissues, including the skin, may take up a substantial portion of their retinol from chylomicrons (59). The lack of this supply in VAD-treated mice provides a possible explanation for their wound healing abnormalities.
The delay in wound closure in VAD-treated mice was mainly a consequence of impaired directional migration of keratinocytes, which may result from alterations in the contact with the underlying matrix. Consistent with this assumption and despite the systemic VAD treatment, proteomics analysis of wound tissue identified fibroblast-enriched proteins as the primary hits. This result points to alterations in fibroblasts and differences in the composition of the granulation tissue as major drivers of the healing defect in VAD-treated mice. In a recent study, VAD treatment of adult mice affected hair follicle stem cell plasticity, which was associated with delayed reformation of the epidermal barrier after wounding (60). Although wound closure, reepithelialization, and granulation tissue formation were not analyzed in that study, the data suggest an additional cell-autonomous effect of retinol deficiency in keratinocytes on wound reepithelialization.
Among the significantly less abundant proteins in wounds of VAD-treated mice was Itga11, a collagen receptor involved in migration and ECM reorganization (61, 62). Upregulation of ITGA11 expression increased migration of cancer-associated fibroblasts, while reduced expression of this gene in dermal fibroblasts impaired their migratory capacity (63, 64). Itga11 expression was induced by mechanosignaling in the skin and is important for myofibroblast differentiation in skin and liver (65, 66). Most importantly, wounds in Itga11-knockout mice had less granulation tissue, lacked myofibroblasts, and showed reduced contractility and tensile strength (41). These findings, together with the reduced levels of Itga11 and α-SMA in wounds of VAD-treated mice, support the notion that myofibroblast differentiation is impaired in wounds of VAD-treated mice. The resulting alterations in the granulation tissue provide a likely explanation for the impaired directional migration of keratinocytes.
Ggcx, a vitamin K-dependent γ-glutamyl carboxylase, was also less abundant in wounds of VAD-treated mice. Gamma-carboxylation is an important protein modification that activates specific proteins (67). Its reduction might affect periostin, which requires γ-carboxylation in some, but not all tissues (68, 69). In skin wounds, periostin facilitates cell-matrix interactions by binding to αVβ3 and αVβ5 integrins and modulates myofibroblast differentiation (43, 70).
In conclusion, we applied multiple omics technologies to identify important pathways in wound fibroblasts. We also established a targeted proteomics strategy that allows the identification of multiple components of the retinol metabolism pathway in any tissue. Therefore, our work provides important datasets and tools for the study of wound healing and retinol metabolism. Most importantly, we identified retinol metabolism as a key pathway in wound healing, which opens avenues for its precise manipulation for the treatment for impaired healing.
-
Methods
Sex as a biological variable
The wound healing studies were only performed with female mice, because male mice often have bite wounds and scars that may affect the healing process and because previous studies showed similar effects of retinoids on wound healing in male and female rodents and humans. While the overall mechanisms of wound healing are comparable between sexes, differences in RA metabolism between female and male mice during wound healing cannot be excluded.
Mice
RNA-Seq and metabolomics experiments were performed with skin and wound tissue from wild-type mice in CD1 genetic background. Stra6-KO mice (35) and wild-type C57BL/6 mice of the same breedings were used for wound healing studies. For VAD experiments, wild-type C57BL/6 mice were either from in-house breeding or purchased from Elevage Janvier. Mice were housed under specific pathogen–free conditions in a 12-hour dark/12-hour light cycle. They received water and food ad libitum.
Mouse genotyping
Genomic DNA was isolated from biopsies obtained from ear clipping (16). Stra6 alleles were identified as described (35). Primers are detailed in Supplemental Table 2.
VAD
VAD was started at the age of 4 weeks until the wound healing experiment was performed at the age of 10 weeks. The food retinol content was guaranteed to be lower than 0.22 IU/g according to the manufacturer (Hespra Bio and Kliba Nafag). Control animals received a normal chow diet with the same ingredients but containing 23.1 IU/g retinol.
Wound healing experiments
Wound healing experiments were performed as previously described (16). Wounds were collected using a 5 mm punch biopsy, and the subcutaneous fat was removed. For stainings, wounds were bisected to obtain sections from the middle of the wound.
FACS isolation of skin and wound fibroblasts
FACS isolation of fibroblasts, including the gating strategy, was previously described (16). Between 80,000 and 100,000 cells were sorted. For metabolomics, they were washed with ammonium carbonate wash solution (75 mM ammonium carbonate in H2O, pH 7.4, freshly prepared) and centrifuged at 1,000g for 10 minutes. The supernatant was removed, and the cells were snap-frozen in liquid nitrogen and stored at –80°C.
RNA isolation and RT-qPCR from tissues
Total RNA was isolated from shock-frozen tissue pieces using TRIzol. Reverse transcription was performed with 500–1,000 μg of RNA using the iScript cDNA synthesis kit (Bio-Rad). RT-qPCR (16) was performed using primers detailed in Supplemental Tables 4 and 5.
Western blot analysis
RIPA buffer (300 μL) was added to snap-frozen tissue samples. The tissue was disrupted with a TissueLyser II with shaking at 30 Hz for 5 minutes. Protein concentration was determined using the BCA assay (Thermo Fisher Scientific). Protein concentrations were adjusted with RIPA buffer, and samples were denatured by addition of Laemmli buffer and incubation at 95°C for 5 minutes. Proteins were separated by polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Unspecific binding sites were blocked by incubation in 5% skim milk in TBS with Tween 20 for 1 hour, and the membranes were incubated with the primary antibody overnight (o/n) at 4°C. After washing, HRP-coupled secondary antibodies were added for 1 hour at room temperature (RT). Antibodies used are shown in Supplemental Table 3.
Metabolomics
FACS-sorted cells were stored at –80°C and extracted with 50 μL cold (–20°C) 40:40:20 acetonitrile/methanol/water. After 10 minutes of incubation at –20°C with sporadic vortexing, cells were pelleted by centrifugation at 4°C and full speed in a table-top centrifuge. The clear supernatant (20–30 μL) was transferred to a plate for injection into the mass spectrometer. Untargeted metabolomics by flow injection mass spectrometry (MS) was performed as described (71). The samples were injected at an isocratic flow rate of 150 μL/min of isopropanol/H2O (6:4, v/v) containing ammonium fluoride (1 mM) and the reference compounds hexakis (2,2,3,3-tetrafluoropropoxy)phosphazene and homotaurine (3-amino-1-propane sulfonic acid). Electrospray ionization was used with the following source parameters: gas temperature 225°C, drying gas 11 L/min, nebulizer 20 psi, sheath gas temperature 350°C, sheath gas flow 10 L/min, Vcap 3,500 V, and nozzle voltage 2,000 V. The fragmentor was set at 350 V, and the Oct1RF Vpp was set at 650 V. The mass spectrometer was operated in full scan mode scanning the mass range (50–1,000 m/z) at 1.4 spectra per second. An online mass correction was performed using the reference masses 138.0230374 and 940.0003763.
Raw MS data were converted to an open-source format (mz5) and processed using an in-house data processing pipeline (FiaMiner) in Matlab (MATHWORKS). The m/z axis for the whole dataset was recalibrated using expected masses. Afterward, annotation was performed based on m/z, with mass accuracy of 0.001 Da matching to the human metabolome database. Differential analysis statistics was performed in Matlab for pairwise comparisons followed by correction of multiple comparisons.
Determination of retinoid content by targeted MS
Liquid-liquid extraction. All procedures were performed in the dark. Samples were thawed on ice and briefly vortexed. Protein content of the 1:100 diluted samples was measured using a Lunatic spectrophotometer (Unchained Labs). A total of 70 μL of each sample was transferred to a glass tube, 7 μL of 1.1 M HCl and 578 μL of methanol (MeOH) were added and vortexed, 1,925 μL of methyl tert-butyl ether were added, and the mixture was vortexed again and incubated for 15 minutes at 25°C in a sonication bath. Then 481 μL of Milli-Q water (MilliporeSigma) were added, vortexed, and centrifuged for 5 minutes at 6,250g and 4°C. The upper organic phase was collected and dried under a nitrogen stream at 30°C. The aqueous phase and pellets were stored at –80°C. The evaporation residues of the organic phase were reconstituted in 105 μL 75% MeOH/0.1% formic acid, vortexed, and subjected to shaking at 1,400 rpm for 15 minutes at 15°C, before being vortexed again. The solution was centrifuged at 18,500g for 10 minutes at 4°C, and the supernatant was analyzed by liquid chromatography-tandem mass spectrometry on a triple-quadrupole mass spectrometer using selected reaction monitoring (for details see Supplemental Tables 6 and 7).
Lipidomics
For quantitative lipidomics, internal standards were added prior to lipid extraction. Tissue samples of 2 mg wet weight were subjected to lipid extraction (72). Lipid analysis was performed by direct flow injection analysis (FIA) using a triple-quadrupole mass spectrometer (FIA-MS/MS, Quattro Ultima) and a high-resolution hybrid quadrupole-Orbitrap mass spectrometer (FIA-FTMS, Thermo Fisher Scientific). FIA-MS/MS was performed in positive ion mode using the analytical setup and strategy described previously (73). A fragment ion of m/z 184 was used for lysophosphatidylcholines (74). The following neutral losses were applied: phosphatidylethanolamine (PE) and lysophosphatidylethanolamine 141, phosphatidylserine 185, phosphatidylglycerol (PG) 189, and phosphatidylinositol 277 (75). Sphingosine-based ceramides and hexosylceramides were analyzed using a fragment ion of m/z 264 (76). PE-based plasmalogens were analyzed according to the principles described by Zemski Berry (77). Cardiolipin was monitored by diglycerol fragment ions (78). Glycerophospholipid species annotation was based on the assumption of even-numbered carbon chains only.
FIA-FTMS was performed as described (79). Triglycerides, diglycerides, and cholesterol esters (CE) were recorded in positive-ion mode m/z 500–1,000 as [M+NH4]+ at a target resolution of 140,000 (at 200 m/z). CE species were corrected for their species-specific response (80). Phosphatidylcholines (PC), PC ether, and sphingomyelins were analyzed in negative ion mode m/z 520–960 as [M+HCOO]– at the same resolution setting. Analysis of free cholesterol (FC) was performed by multiplexed acquisition of the [M+NH4]+ of FC and the deuterated internal standard (FC[D7]) (80). Free fatty acids were analyzed in negative ion mode m/z 150–450 as [M-H]– dissolved in MeOH/chloroform 5:1 (v/v) containing 0.005% dimethylamine.
Bioinformatic analysis of RNA-Seq and metabolomics data
Overrepresentation analysis. Overrepresentation analysis was performed using ClusterProfiler in RStudio 2023.06.0+421 (R version 4.3.2) (81). Differentially expressed genes were defined by a |log2(fold change)| over 0.5 and a P value below 0.05.
Integrated pathway analysis. A joint pathway analysis was performed using MetaboAnalyst 6.0 (82). Significantly differentially expressed genes (P ≤ 0.05) and significantly differentially abundant metabolites (P ≤ 0.05) found in both experiments were used for the analysis.
Bioinformatic analysis of scRNA-Seq data sets
The scRNA-Seq data sets from mouse skin wounds were downloaded from Gene Expression Omnibus (GEO) (GSE204777) (26) and reanalyzed using the Seurat R package (version 4.2.0). The data from the 5 batches were first analyzed separately for quality control by eliminating low-quality cells with low RNA features, as well as those with relatively high mitochondrial and ribosomal RNA content via the subset function, followed by SCtransform using the vars.to.regress modifier to additionally regress out the mitochondrial and ribosomal RNA features (83). Time point– and wound location–specific barcodes were determined for each cell using the HTODemux hash tag oligo demultiplexing function. The data sets were merged and integrated using the merge, SelectIntegrationFeatures, PrepSCTIntegration, FindIntegrationAnchors and IntegrateData functions, according to the published data analysis and integration protocol (84). The integrated data sets were analyzed via the RunPCA function, followed by ElbowPlot to visualize the standard deviation of the principal components and to select the most significant dimensions for further analyses. The RunUMAP and FindNeighbors functions were run using standard settings and the dims=1:30 modifier, followed by FindClusters with resolution=0.1 to cluster the data set on major cell types. Marker genes of skin cells were obtained from CellMarker 2.0 (85) and mapped onto the data set by DotPlot and FeaturePlot. Immune cells, fibroblasts, contractile cells, epithelial cells, endothelial cells, and others were identified and annotated. Using additional cell markers, the following subtypes of the major cell types were identified and annotated: neutrophils, macrophages, monocytes, early fibroblasts, T cells, activated fibroblasts, late fibroblasts, basal keratinocytes, endothelial cells, smooth muscle cells, keratinocytes, melanocytes, and erythrocytes. Wound fibroblasts were analyzed by first subsetting the fibroblast clusters via the Subset function, followed by the downstream steps outlined above. Marker genes for fibroblast subtypes were determined by the FindAllMarkers function. Space/time tile plots for individual genes were generated as described (26).
IHC
FFPE samples were dewaxed, rehydrated, and washed with PBS. Antigen retrieval was performed with 0.1 M sodium citrate buffer (pH 6) at 95°C in a water bath and incubation for 1 hour. After 30 minutes of cooling at RT and washing, slides were incubated in blocking buffer (12% BSA in PBS with Tween 20) for 1 hour at RT. The primary antibody in blocking buffer was incubated at 4°C o/n. After washing, the biotinylated secondary antibody was added in blocking buffer for 1 hour at RT. Developing was performed as described by the manufacturer of the VECTASTAIN Elite ABC-HRP Kit (Vector Laboratories). After washing with water, sections were counterstained with hematoxylin for 3 minutes, dehydrated, and mounted with Mowiol. Antibodies used are specified in Supplemental Table 3.
Quantification of histological stainings. Quantification of Herovici staining was performed using a custom script in ImageJ (NIH) (86). Light blue pixels resulting from young (type III–enriched) collagen fibers stained with methyl blue, while dark purple pixels resulting from matured (type I–enriched) collagen fibers stained with acid fuchsin (87). Areas with these colors of interest were manually selected to define color ranges for later pixel-based analysis, and the granulation tissue was manually segmented. Within the segmented area, the chosen color ranges were used to define pixels positive for young or mature collagen, respectively. The same color segmentation setting was applied to all images. The positively stained pixels were plotted as percentage of total pixels within the total granulation tissue area.
Ki67 staining was quantified using QuPath 0.5.0 (88). Ki67+ cells in the wound epidermis were quantified using the positive cell detection feature of QuPath and then calculating the percentage of positive cells among all cells and the number of positively stained epidermal cells per micrometer or millimeter of epidermis.
Collagenous and noncollagenous peptides in Picrosirius red/fast green FCF-stained sections were quantified using QuPath 0.5.0. Red (collagenous peptides, Picrosirius red) and green (noncollagenous peptides, fast green FCF) pixels were thresholded in the manually segmented granulation tissue, and positively classified areas were measured. Collagen area ratio was calculated by taking the ratio of the Picrosirius red–positive and the fast green–positive area.
MECA-32 and α-SMA stainings were analyzed using QuPath 0.5.0. For quantification of the α-SMA and MECA-32–positive areas, the granulation tissue was manually segmented. The MECA-32– (Cy3) positive area was determined by thresholding and normalized to the granulation tissue area. To establish a threshold for specific signal detection and minimize background interference in the α-SMA staining, the mean gray value and SD of the nonspecific signal seen in the secondary antibody control were quantified. A cutoff value was defined as the mean gray value plus twice the SD (μ + 2σ). Signals in the stained sections exceeding this threshold were considered specific, while those below were attributed to background and excluded from analysis. The α-SMA–positive area was normalized to the granulation tissue area.
Proteomics
Protein lysate preparation. Snap-frozen wound and skin samples were transferred into 400 μL 4 M guanidine hydrochloride in 250 mM HEPES, pH 7.8. Metal beads were added, and the tissues were lysed using a TissueLyser II at 30 Hz in 2 cycles of 2 minutes, followed by 1 cycle of 1 minute. Samples were sonicated for 10 cycles of 30 seconds on/30 seconds off at 4°C and centrifuged at 10,000g for 10 minutes at 4°C. A volume equivalent to 20 μg of protein was diluted to 40 μL with lysis buffer. For this purpose, 1 μL of 350 mM Tris(2-carboxyethyl)phosphine was added to a final concentration of 10 mM, followed by the addition of 2.8 μL of freshly prepared 500 mM chloroacetamide solution to a final concentration of 40 mM. The mixtures were incubated for 5 minutes at 95°C and then diluted to 2 M guanidine hydrochloride with 45 μL of 250 mM HEPES buffer. Subsequently, 0.5 μL of a 1 μg/μL of Lys-C endoproteinase stock solution was added to achieve an enzyme/protein (w/w) ratio of 1:40, and the samples were digested for 3 hours at 37°C while shaking at 350 rpm. They were further diluted to 0.5 M guanidine hydrochloride by adding 240 μL of 250 mM HEPES buffer, before adding 1 μL of a 1 μg/μL trypsin stock solution to achieve an enzyme/protein lysate (w/w) ratio of 1:20. Samples were incubated o/n at 37°C with shaking at 350 rpm. Trypsin was inactivated by acidifying the samples with approximately 90 μL of 10% trifluoroacetic acid (TFA) to achieve a final concentration of around 1% TFA and a pH of 1–2.
EvoTip preparation and sample loading. We added 20 μL of acetonitrile to each EvoTip column, followed by centrifugation at 700g for 1 minute. Tips were soaked in 100% isopropanol for 20 seconds. While soaking, 20 μL of 0.1% formic acid (FA) was added to the top of the tips, and the solution was removed by centrifugation at 700g for 1 minute. A sample containing 750 ng of protein was diluted to a final volume of 20 μL with 0.1% FA and added to the tips. The tips were centrifuged at 700g for 1 minute, followed by 2 washes with 20 μL 0.1% FA, each step including centrifugation at 700g for 1 minute. After the second wash, 250 μL of 0.1% FA was added to the top and centrifuged at 700g for 10 seconds. The loaded tips were stored soaked in 0.1% FA until analysis.
DIA data acquisition. The analysis was conducted using an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific) in combination with an LC Evosep One system. Samples were run using the pre-programmed Whisper100 10SPD method on an Acclaim PepMap RSLC C18 column (1.9 μm, 75 μm × 150 mm, Thermo Fisher Scientific). The mass spectrometer operated in positive ion mode and was coupled with a FAIMS Pro interface, with the compensation voltage set to –45 V. MS scans were recorded at a resolution of 120,000, covering a mass range of 400–1,000 m/z, with an AGC target of 300% and maximum injection time set to auto. The DIA isolation windows also spanned 400–1,000 m/z. Precursor ions were fragmented using higher-energy collisional dissociation (HCD) with a normalized collision energy of 32%. Fragment ion scans were acquired at a resolution of 60,000, with an automatic gain control (AGC) target of 1,000% and the maximum injection time set to auto.
DIA data analysis. Raw DIA data files were analyzed using Spectronaut 17 (Biognosys) in DirectDIA (Deep) mode, utilizing the mouse UniProt database (taxonomy ID: 10090) with modified Biognosys factory settings. The enzyme specificity was set for database searches to trypsin and Lys-C, allowing up to 1 missed cleavage. Carbamidomethylation of cysteine was selected as a fixed modification, while acetylation of the protein N-terminus and methionine oxidation were considered variable modifications. Quantification was performed at the MS1 level, focusing exclusively on proteotypic peptides. Protein quantification data were extracted and subjected to further analysis using a custom Python script (Python version 3.11.4), which also performed the 2-way ANOVA for statistical assessment.
SureQuant analysis. Three peptides per protein, which identify both human and mouse orthologs of the proteins (Supplemental Table 8), were chosen for assay validation and suitability. Human reference peptides were taken from Proteomics DB (https://www.proteomicsdb.org/proteomicsdb/#human), and mouse reference peptides were identified from https://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msdigest Whenever possible, 3 peptides present in both reference peptide sets were used. The final selection of peptides was based on a defined set of criteria, including frequent and high-confidence identification in MS experiments. The peptides were required to meet specific conditions: absence of methionine, a length between 8 and 21 amino acids, a C-terminal arginine or lysine, and no missed cleavages, whenever possible. The heavy isotope–labeled arginine or lysine peptides, used for SureQuant analysis, were purchased from JPT Peptide Technologies.
SureQuant data acquisition. SureQuant analysis was performed on an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific) in combination with an LC Evosep One system. Samples were run using the pre-programmed 88-minute gradient on an Acclaim PepMap RSLC C18 column (1.9 μm, 75 μm × 150 mm, Thermo Fisher Scientific). A total of 750 ng of sample along with 500 fmol of heavy peptides was used for the analysis. The MS1 scan was performed with a resolution of 120,000, an AGC target of 300%, an injection time of 50 ms, and a cycle time of 3 seconds. This was followed by the detection of heavy peptide precursors and their fragments based on a predefined list (selected proteins and respective peptides are detailed in Supplemental Table 8), using a resolution of 7,500, HCD energy of 30, AGC target of 1,000%, and a 10 ms injection time. The product ion trigger required detection of at least 2 product ions to initiate an offset scan, which was carried out at a resolution of 120,000, HCD energy of 30, AGC target of 1,000%, and a 250 ms injection time in profile mode.
SureQuant data analysis. The SureQuant data were analyzed using Skyline version 23.1. Target peptides were imported with the MS1 filter parameters set to centroided precursor mass analyzer, mass accuracy of 10 ppm, and MS/MS filter parameters set to SureQuant acquisition method, with Orbitrap as a product mass analyzer at a resolution of 60,000 at 200 m/z. Each precursor mass was configured to monitor at least 3 product ions. All peptides included a fixed modification of carbamidomethylation on cysteine (+57.021 Da) and oxidation on methionine (+15.994 Da). Synthetic peptides also had static modifications with heavy isotopes, lysine (+8.014 Da) and arginine (+10.008 Da). Peptide transition peaks were reviewed and adjusted manually using the retention times of the heavy peptides as references. Peptide fragment areas were normalized to the total ion current chromatogram area. The total area fragment for each precursor was used for quantification. Protein quantification was performed by averaging the values of its detected peptides.
Statistics
All statistical data are based on biological replicates. Statistical analysis was performed with Prism software, version 10 for Windows (GraphPad Software). Mann-Whitney U test was used for the comparison of 2 groups. For comparisons involving more than 2 groups, we used Šídák’s or Tukey’s multiple comparisons test as specified in the figure legends. A P value of less than 0.05 was considered significant.
Study approval
Mouse experiments and maintenance of mice were approved by the local veterinary authorities (Kantonales Veterinäramt Zürich, Zurich, Switzerland).
Data availability
RNA-Seq data (16) had been deposited at GEO. They are available under the accession number GSE134789.
The proteomics data had been submitted to the ProteomeXchange Consortium via the PRIDE repository and are available under the accession codes PXD061167 (DIA dataset) and PXD061256 (SureQuant dataset) (89, 90).
Raw data of the metabolomics analysis had been uploaded to MassIVE and are available under accession number MSV000097301 (ftp://massive.ucsd.edu/v09/MSV000097301/).
All original data used to create the figures of this manuscript are available in the Supporting Data Values file.
Further information can be found in Supplemental Methods.
-
Author contributions
TW designed and performed all cell culture and wound healing experiments, designed the proteomics experiment with EM and UADK, conducted the proteomics experiment and analysis with EM, performed the integration of the bulk RNA-Seq and metabolomics experiments, and wrote the manuscript together with SW. SW led the study from conception to design of experiments, provided troubleshooting, and supported analysis of the data. EM performed the DIA analysis. MSW performed the FACS experiment and reanalyzed published scRNA-Seq data. NBG provided Stra6-KO mice and expertise in retinoid biology. MH and GL designed, performed, and analyzed the lipidomics experiments. NZ designed and performed the metabolomics experiment. All authors provided important feedback to the manuscript.
-
Funding support
Swiss National Science Foundation (310030-212212 to SW).
-
Acknowledgments
We thank Sol Dominic Taguinod and other staff of the ETH Zurich Phenomics Center for mouse colony maintenance; Drs. Alaa Othman and Stefan Schauer, Functional Genomics Center Zurich, for targeted retinol measurements; and Dr. Hans-Dietmar Beer, University Hospital Zurich, for human primary fibroblasts. SW, NZ, and UADK are/were members of the SKINTEGRITY. CH collaborative research consortium. TW was a member of the Molecular Life Sciences PhD program of ETH Zurich and the University of Zurich.
Address correspondence to: Sabine Werner, Institute of Molecular Health Sciences, ETH Zurich, Otto-Stern-Weg 7, 8093 Zürich, Switzerland. Email: sabine.werner@biol.ethz.ch.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2025, Wüstemann et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2025;10(22):e194188.https://doi.org/10.1172/jci.insight.194188.
-
References
- Martin P. Wound healing--aiming for perfect skin regeneration. Science. 1997;276(5309):75–81.
- Gurtner GC, et al. Wound repair and regeneration. Nature. 2008;453(7193):314–321.
- Foster DS, et al. Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proc Natl Acad Sci U S A. 2021;118(41):e2110025118.
- Driskell RR, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504(7479):277–281.
- Rinkevich Y, et al. Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science. 2015;348(6232):aaa2151.
- Plikus MV, et al. Regeneration of fat cells from myofibroblasts during wound healing. Science. 2017;355(6326):748–752.
- Bergmeier V, et al. Identification of a myofibroblast-specific expression signature in skin wounds. Matrix Biol. 2018;65:59–74.
- Shook BA, et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science. 2018;362(6417):eaar2971.
- Guerrero-Juarez CF, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun. 2019;10(1):650.
- Eming SA, et al. Metabolic orchestration of the wound healing response. Cell Metab. 2021;33(9):1726–1743.
- Peña OA, Martin P. Cellular and molecular mechanisms of skin wound healing. Nat Rev Mol Cell Biol. 2024;25(8):599–616.
- Manchanda M, et al. Metabolic reprogramming and reliance in human skin wound healing. J Invest Dermatol. 2023;143(10):2039–2051.
- Willenborg S, et al. Mitochondrial metabolism coordinates stage-specific repair processes in macrophages during wound healing. Cell Metab. 2021;33(12):2398–2414.
- Sood RF, et al. Targeted metabolic profiling of wounds in diabetic and nondiabetic mice. Wound Repair Regen. 2015;23(3):423–434.
- Zhao X, et al. Metabolic regulation of dermal fibroblasts contributes to skin extracellular matrix homeostasis and fibrosis. Nat Metab. 2019;1(1):147–157.
- Wietecha MS, et al. Activin-mediated alterations of the fibroblast transcriptome and matrisome control the biomechanical properties of skin wounds. Nat Commun. 2020;11(1):2604.
- Collins CA, et al. Reprogramming adult dermis to a neonatal state through epidermal activation of β-catenin. Development. 2011;138(23):5189–5199.
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83(3):835–870.
- Li Y-T, et al. PDGFRα-lineage origin directs monocytes to trafficking proficiency to support peripheral immunity. Eur J Immunol. 2022;52(2):204–221.
- Binek A, et al. Flow cytometry has a significant impact on the cellular metabolome. J Proteome Res. 2019;18(1):169–181.
- Li Y, et al. The multifaceted nature of retinoid transport and metabolism. Hepatobiliary Surg Nutr. 2014;3(3):126–139.
- Amengual J, et al. Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J Biol Chem. 2012;287(29):24216–24227.
- Kim Y-K, et al. Retinyl ester formation by lecithin: retinol acyltransferase is a key regulator of retinoid homeostasis in mouse embryogenesis. J Biol Chem. 2008;283(9):5611–5621.
- Kawaguchi R, et al. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315(5813):820–825.
- Napoli JL. Retinol metabolism in LLC-PK1 Cells. Characterization of retinoic acid synthesis by an established mammalian cell line. J Biol Chem. 1986;261(29):13592–13597.
- Hu KH, et al. Transcriptional space-time mapping identifies concerted immune and stromal cell patterns and gene programs in wound healing and cancer. Cell Stem Cell. 2023;30(6):885–903.
- Iyer VR, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283(5398):83–87.
- Obrochta KM, et al. Effects of diet and strain on mouse serum and tissue retinoid concentrations. PLoS One. 2014;9(6):e99435.
- Sommer A, Davidson FR. Assessment and control of vitamin A deficiency: the Annecy Accords. J Nutr. 2002;132(9 suppl):2845S–2850S.
- Al Tanoury Z, et al. Genes involved in cell adhesion and signaling: a new repertoire of retinoic acid receptor target genes in mouse embryonic fibroblasts. J Cell Sci. 2014;127(pt 3):521–533.
- Delacroix L, et al. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30(1):231–244.
- Klein ES, et al. Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J Biol Chem. 1996;271(37):22692–22696.
- Hübner G, et al. Differential regulation of pro-inflammatory cytokines during wound healing in normal and glucocorticoid-treated mice. Cytokine. 1996;8(7):548–556.
- auf dem Keller U, et al. Reactive oxygen species and their detoxification in healing skin wounds. J Investig Dermatol Symp Proc. 2006;11(1):106–111.
- Berry DC, et al. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J Biol Chem. 2013;288(34):24528–24539.
- Berry DC, et al. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc Natl Acad Sci U S A. 2011;108(11):4340–4345.
- Kato-Suzuki M, et al. Time-dependent changes in retinoids content in liver and adipose tissue after feeding of a vitamin A-deficient diet to mice. Exp Anim. 2024;73(3):302–309.
- Relevy NZ, et al. Vitamin A-deficient diet accelerated atherogenesis in apolipoprotein E(−/−) mice and dietary β-carotene prevents this consequence. BioMed Res Int. 2015;2015:758723.
- Chen L, et al. The murine excisional wound model: contraction revisited. Wound Repair Regen. 2015;23(6):874–877.
- Igata T, et al. Loss of the transcription repressor ZHX3 induces senescence-associated gene expression and mitochondrial-nucleolar activation. PLoS One. 2022;17(1):e0262488.
- Schulz J-N, et al. Reduced granulation tissue and wound strength in the absence of α11β1 integrin. J Invest Dermatol. 2015;135(5):1435–1444.
- Shiba S, et al. γ-Glutamyl carboxylase in osteoblasts regulates glucose metabolism in mice. Biochem Biophys Res Commun. 2014;453(3):350–355.
- Ontsuka K, et al. Periostin, a matricellular protein, accelerates cutaneous wound repair by activating dermal fibroblasts. Exp Dermatol. 2012;21(5):331–336.
- Abbasi S, et al. Distinct regulatory programs control the latent regenerative potential of dermal fibroblasts during wound healing. Cell Stem Cell. 2020;27(3):396–412.
- Sinha S, et al. Fibroblast inflammatory priming determines regenerative versus fibrotic skin repair in reindeer. Cell. 2022;185(25):4717–4736.
- Podolak-Popinigis J, et al. Transcriptome profiling reveals distinctive traits of retinol metabolism and neonatal parallels in the MRL/MpJ mouse. BMC Genomics. 2015;16(1):926.
- Szebeni A, et al. [Vitamin A serum levels in burned patients]. Orv Hetil. 1981;120(40):2413–2415.View this article via: PubMed Google Scholar
- Rojas AI, Phillips TJ. Patients with chronic leg ulcers show diminished levels of vitamins A and E, carotenes, and zinc. Dermatol Surg. 1999;25(8):601–604.
- Polcz ME, Barbul A. The role of vitamin A in wound healing. Nutr Clin Pract. 2019;34(5):695–700.
- Ehrlich HP, Hunt TK. Effects of cortisone and vitamin A on wound healing. Ann Surg. 1968;167(3):324–328.
- Kitano Y, et al. Pretreatment with topical all-trans-retinoic acid is beneficial for wound healing in genetically diabetic mice. Arch Dermatol Res. 2001;293(10):515–521.
- de Campos Peseto D, et al. Effects of tretinoin on wound healing in aged skin. Wound Repair Regen. 2016;24(2):411–417.
- Hung VC, et al. Topical tretinoin and epithelial wound healing. Arch Dermatol. 1989;125(1):65–69.
- Paquette D, et al. Short-contact topical tretinoin therapy to stimulate granulation tissue in chronic wounds. J Am Acad Dermatol. 2001;45(3):382–386.
- Correa-Gallegos D, et al. CD201+ fascia progenitors choreograph injury repair. Nature. 2023;623(7988):792–802.
- Griffin M, et al. Multi-omic analysis reveals retinoic acid molecular drivers for dermal fibrosis and regenerative repair in the skin. Cell Stem Cell. 2025;32(9):1421–1437.
- Chassaing N, et al. Phenotypic spectrum of STRA6 mutations: from Matthew-Wood syndrome to non-lethal anophthalmia. Hum Mutat. 2009;30(5):E673–E681.
- Golzio C, et al. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am J Hum Genet. 2007;80(6):1179–1187.
- Hodges JK, et al. Vitamin A supplementation increases the uptake of chylomicron retinyl esters into the brain of neonatal rats raised under vitamin A-marginal conditions. J Nutr. 2016;146(9):1677–1683.
- Tierney MT, et al. Vitamin A resolves lineage plasticity to orchestrate stem cell lineage choices. Science. 2024;383(6687):eadi7342.
- Tiger C-F, et al. Alpha11beta1 integrin is a receptor for interstitial collagens involved in cell migration and collagen reorganization on mesenchymal nonmuscle cells. Dev Biol. 2001;237(1):116–129.
- Alam J, et al. Generation of a novel mouse strain with fibroblast-specific expression of Cre recombinase. Matrix Biol Plus. 2020;8:100045.
- Zhou P, et al. KRT17 from keratinocytes with high glucose stimulation inhibit dermal fibroblasts migration through integrin α11. J Endocr Soc. 2024;8(2):bvad176.
- Iwai M, et al. Cancer-associated fibroblast migration in non-small cell lung cancers is modulated by increased integrin α11 expression. Mol Oncol. 2021;15(5):1507–1527.
- Bansal R, et al. Integrin alpha 11 in the regulation of the myofibroblast phenotype: implications for fibrotic diseases. Exp Mol Med. 2017;49(11):e396.
- Carracedo S, et al. The fibroblast integrin alpha11beta1 is induced in a mechanosensitive manner involving activin A and regulates myofibroblast differentiation. J Biol Chem. 2010;285(14):10434–10445.
- Dowd P, et al. The mechanism of action of vitamin K. Annu Rev Nutr. 1995;15:419–440.
- Coutu DL, et al. Periostin, a member of a novel family of vitamin K-dependent proteins, is expressed by mesenchymal stromal cells. J Biol Chem. 2008;283(26):17991–18001.
- Annis DS, et al. Absence of vitamin K-dependent γ-carboxylation in human periostin extracted from fibrotic lung or secreted from a cell line engineered to optimize γ-carboxylation. PLoS One. 2015;10(8):e0135374.
- Elliott CG, et al. Periostin modulates myofibroblast differentiation during full-thickness cutaneous wound repair. J Cell Sci. 2012;125(pt 1):121–132.
- Fuhrer T, et al. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal Chem. 2011;83(18):7074–7080.
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917.
- Liebisch G, et al. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim Biophys Acta. 2004;1686(1–2):108–117.
- Liebisch G, et al. High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin Chem. 2002;48(12):2217–2224.
- Matyash V, et al. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49(5):1137–1146.
- Liebisch G, et al. Quantitative measurement of different ceramide species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI-MS/MS). J Lipid Res. 1999;40(8):1539–1546.
- Zemski Berry KA, Murphy RC. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J Am Soc Mass Spectrom. 2004;15(10):1499–1508.
- Scherer M, et al. Simultaneous quantification of cardiolipin, bis(monoacylglycero)phosphate and their precursors by hydrophilic interaction LC-MS/MS including correction of isotopic overlap. Anal Chem. 2010;82(21):8794–8799.
- Höring M, et al. Accurate quantification of lipid species affected by isobaric overlap in Fourier-transform mass spectrometry. J Lipid Res. 2021;62:100050.
- Höring M, et al. Quantification of cholesterol and cholesteryl ester by direct flow injection high-resolution fourier transform mass spectrometry utilizing species-specific response factors. Anal Chem. 2019;91(5):3459–3466.
- Xu S, et al. Using clusterProfiler to characterize multiomics data. Nat Protoc. 2024;19(11):3292–3320.
- Pang Z, et al. MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024;52(w1):W398–W406.
- Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20(1):296.
- Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.
- Hu C, et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2022;51(d1):D870–D876.
- Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682.
- Herovici C. A polychrome stain for differentiating precollagen from collagen. Stain Technol. 1963;38:204–205.View this article via: PubMed Google Scholar
- Bankhead P, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878.
- Deutsch EW, et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2022;51(d1):D1539–D1548.
- Perez-Riverol Y, et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2024;53(d1):D543–D553.
-
Version history
- Version 1 (October 16, 2025): In-Press Preview
- Version 2 (November 24, 2025): Electronic publication










