Research ArticleImmunologyInfectious disease Free access | 10.1172/jci.insight.151782
Cross-reactive antibodies facilitate innate sensing of dengue and Zika viruses
Laura K. Aisenberg,1 Kimberly E. Rousseau,1 Katherine Cascino,1 Guido Massaccesi,1 William H. Aisenberg,2 Wensheng Luo,3 Kar Muthumani,4 David B. Weiner,4 Stephen S. Whitehead,5 Michael A. Chattergoon,1 Anna P. Durbin,3 and Andrea L. Cox1
1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by Aisenberg, L. in: PubMed | Google Scholar
1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Rousseau, K.
in:
PubMed
|
Google Scholar
|

1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Cascino, K.
in:
PubMed
|
Google Scholar
|

1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Massaccesi, G.
in:
PubMed
|
Google Scholar
|

1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Aisenberg, W.
in:
PubMed
|
Google Scholar
|

1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by Luo, W. in: PubMed | Google Scholar
1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Muthumani, K.
in:
PubMed
|
Google Scholar
|

1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Weiner, D.
in:
PubMed
|
Google Scholar
|

1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by Whitehead, S. in: PubMed | Google Scholar
1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by Chattergoon, M. in: PubMed | Google Scholar
1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Durbin, A.
in:
PubMed
|
Google Scholar
|

1Department of Medicine, Division of Infectious Disease, and
2Department of Medicine, Division of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
3Center for Immunization Research, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
4The Wistar Institute, Vaccine & Immunotherapy Center, Philadelphia, Pennsylvania, USA.
5National Institute of Allergy and Infectious Diseases, NIH, Laboratory of Viral Diseases, Bethesda, Maryland, USA.
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
Authorship note: LKA and KER are co–first authors.
Find articles by
Cox, A.
in:
PubMed
|
Google Scholar
|

Authorship note: LKA and KER are co–first authors.
Published May 19, 2022 - More info
JCI Insight. 2022;7(12):e151782. https://doi.org/10.1172/jci.insight.151782.
© 2022 American Society for Clinical Investigation
Received: June 9, 2021; Accepted: May 13, 2022
-
Abstract
The Aedes aegypti mosquito transmits both dengue virus (DENV) and Zika virus (ZIKV) . Individuals in endemic areas are at risk for infection with both viruses, as well as for repeated DENV infection. In the presence of anti-DENV antibodies, outcomes of secondary DENV infection range from mild to life threatening. Furthermore, the role of cross-reactive antibodies on the course of ZIKV infection remains unclear. We assessed the ability of cross-reactive DENV mAbs or polyclonal immunoglobulin isolated after DENV vaccination to upregulate type I IFN production by plasmacytoid DCs (pDCs) in response to both heterotypic DENV- and ZIKV-infected cells. We found a range in the ability of antibodies to increase pDC IFN production and a positive correlation between IFN production and the ability of an antibody to bind to the infected cell surface. Engagement of Fc receptors on the pDC and engagement of epitope on the infected cell by the Fab portion of the same antibody molecule was required to mediate increased IFN production by providing specificity to and promoting pDC sensing of DENV or ZIKV. This represents a mechanism independent of neutralization by which preexisting cross-reactive DENV antibodies could protect a subset of individuals from severe outcomes during secondary heterotypic DENV or ZIKV infection.
-
Introduction
Dengue virus (DENV) and Zika virus (ZIKV) are viral diseases transmitted by the Aedes aegypti mosquito (1, 2). DENV is a rapidly emerging, mosquito-borne viral infection, with an estimated 400 million infections occurring annually. ZIKV is linked to an increased risk of neurologic complications in adults and complications in pregnancy that include microcephaly, preterm birth, and miscarriage (3, 4). Both viruses are members of the Flaviviridae family, genus flavivirus, with highly conserved structural protein identity across them (5–7). While DENV infections can be caused by 4 distinct serotypes of virus, ZIKV has been shown to only exist as a single serotype (8). The role of antibodies in DENV outcomes, particularly those following a secondary infection, is complex. Primary DENV infection generates an antibody response that generally protects against a subsequent symptomatic homotypic infection but does not always provide protection against heterotypic infection. In fact, serotype cross-reactive antibodies can pose a risk for clinically worsened infection (9–13). Why some heterotypic infections result in severe disease while the majority do not remains incompletely understood.
Anti-DENV mAbs can be cross-reactive with ZIKV, which has raised the additional question of how prior exposure to DENV might impact ZIKV infection. Prior studies have investigated the potential impact of ZIKV cross-reactive anti-DENV mAbs on ZIKV infection outcome (14–17). Specifically, data from in vitro models and immunodeficient mouse models have shown that anti-DENV antibodies can increase ZIKV replication (14, 18, 19). However, nonhuman primate (NHP) models and human clinical data are not consistent with these findings (20–27). NHP models demonstrate no worsened clinical outcomes with preexisting DENV antibodies, with some studies demonstrating shortened clinical ZIKV infection after prior DENV exposure (21, 22, 25). Epidemiologic data and analysis of infected people suggest that anti-DENV antibodies may protect against ZIKV infection (24, 26–28).
The type I IFN response is critical to viral control. It has been shown to be directly restrictive of DENV (29–31) and ZIKV (32) replication and to induce a cytokine milieu conducive to generating a subsequent adaptive immune response mediating viral control (33). Both viruses have evolved multiple mechanisms to evade the IFN pathway (34–40), demonstrating the importance of the selective pressure of IFN-mediated restriction of viral replication. Specifically, there is extensive literature demonstrating that the DENV NS5 protein inhibits STAT2 and subsequent IFN production (35, 36). Additional work suggests that DENV subgenomic RNA inhibits TRIM25, further limiting the production of type I IFNs (38). ZIKV has also developed several similar mechanisms, including blocking STAT2 activity via NS5 (39, 40). Additional mechanisms by which ZIKV evades the IFN pathway include inhibition of TANK-binding kinase 1 (TBK1) (41), cleavage of cyclic-GMP-AMP synthase (cGAS) (42), and blockade of the IFN promoter (43). Consistent with an essential role for IFN, individuals with severe cases of dengue fever often have lower IFN levels relative to patients with less severe infections (44–47), and ZIKV-infected fetuses are better protected from severe neurologic sequelae when a robust IFN response is generated (48). Notably, more virulent strains of ZIKV have been demonstrated to induce lower IFN responses than less virulent strains (49).
As in many other viral infections, plasmacytoid DCs (pDCs) are a major producer of type I IFNs in DENV infection (50–54). Prior studies on DENV and ZIKV (55–58) demonstrate that, in order for a type I IFN response to be generated, pDCs require cell-to-cell contact with DENV- or ZIKV-infected cells (59–61). The cell adhesion molecules ICAM-1 (CD54) and αL-integrin are required for the establishment of this cell-to-cell contact (60). Once contact between the pDC and a target cell is established, pathogen-associated molecular patterns (PAMPs) are transferred via an interferogenic synapse from the target cell to pDC, with pDC sensing of viral RNA occurring via TLR7 and subsequent downstream signaling (60).
Despite prior work suggesting that antiviral antibodies can directly alter innate immune signaling pathways (62) and evidence that demonstrates that recent DENV infection offers increased ZIKV control in the setting of increased pDC frequency (63), it is unknown whether antibodies against DENV modulate pDC sensing of either DENV or ZIKV. We sought to determine the effects of anti-DENV antibodies on the production of type I IFN by pDC upon DENV or ZIKV exposure, including whether antibodies generated in DENV infection can upregulate IFN production in response to heterotypic DENV or to ZIKV, potentially increasing protection.
To investigate this hypothesis, we adapted previously defined in vitro models of DENV and ZIKV sensing by pDC (59, 60). With anti-DENV mAbs and unique plasma specimens from DENV-naive human participants challenged with DENV after administration of live attenuated DENV vaccines, we demonstrate that some anti-DENV antibodies facilitate pDC sensing of cross-serotype DENV or of ZIKV and promote type I IFN production. We show that a subset of anti-DENV antibodies function in place of or in concert with cell adhesion molecules to tether DENV- or ZIKV-infected cells to pDCs. This link occurs via binding of Fab to epitopes on infected cells and Fc binding to pDC Fc receptor (FcR), offering a specificity to cell-to-cell contact and generating a more robust antiviral IFN response.
-
Results
pDCs sense DENV in a cell-to-cell contact–dependent manner facilitated by cell adhesion molecules ICAM-1 and αL-integrin. Primary human pDCs produce type I IFN when cocultured with DENV2–New Guinea C–infected (DENV2-NGC–infected) Huh 7.5.1 hepatoma cells. Work by others has demonstrated that pDC sensing of DENV requires direct cell-to-cell contact with infected cells (59, 60). We confirmed that pDC production of IFN is much more robust in the setting of cell-to-cell contact by incubating pDCs for 24 hours with uninfected Huh 7.5.1 cells, with DENV-infected Huh 7.5.1 cells, with DENV alone, with supernatant collected from DENV infected Huh 7.5.1 cells (Figure 1A), or with the TLR7/8 agonist resiquimod (REQ) as a positive control. Supernatant from DENV-infected Huh 7.5.1 cells was titered to confirm the presence of infectious viral particles (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.151782DS1). We then measured IFN-α2a production by ELISA. There is significant donor-to-donor variability between experiments, and it is not feasible to use the same pDC donor for repeated experiments, given the anonymous nature of the leukapheresis donation. Therefore, the amount of IFN generated is reported as a percentage of the pDC response to DENV alone and as the average of at least 3 replicates (Figure 1A). All conditions were tested in the same experiment using the same pDC donor. To demonstrate that pDCs are required for type I IFN production, we also assessed IFN-α2a from DENV infected Huh 7.5.1 cells without pDCs added. We observed that only the pDCs cocultured with infected Huh 7.5.1 cells produced significant amounts of type I IFN, confirming the previous finding that pDCs are required for type I IFN production and that pDCs produce IFN much more robustly when in contact with DENV-infected cells (Figure 1A). Given these data, all subsequent experiments used this coculture model (Figure 1B).
 Figure 1
Figure 1Contact between DENV-infected cells and pDCs is required for type I IFN response. (A) Primary human pDCs were cocultured with uninfected Huh 7.5.1 cells (first bar), with infected Huh 7.5.1 cells (second and third bar), or with DENV alone (fourth bar). Supernatants were collected and IFN-α2a was measured. To assess the requirement for pDCs in IFN production, infected Huh 7.5.1 cells were cultured alone (seventh bar). To assess the requirement for cell-to-cell contact, pDCs were cultured with supernatant collected from infected Huh 7.5.1 cells (fifth bar). To demonstrate that TLR signaling is intact in the pDCs in the absence of hepatoma cells, pDC were cultured with 1 μg/mL TLR7 agonist resiquimod (REQ) (sixth bar). This figure represents 4 independent experiments with distinct pDC donors, with n ≥ 3 per condition. (B) Schematic representation of experimental design to follow: Huh 7.5.1 cells are infected with DENV at 0.1–1 MOI for 48 hours. At 48 hours, antibody treatment is added. pDCs are isolated and cocultured with treated Huh 7.5.1 one hour after antibody addition. Supernatants are collected after 24 hours and analyzed for IFN-α2a. (C and D) Primary human pDCs were cocultured with infected Huh 7.5.1 following 1 hour of treatment with anti–ICAM-1 antibody at 2–10 μg/mL (C) or anti–αL-integrin antibody at 0.1–1 μg/mL (D). Supernatants were collected, and IFN-α2a was measured. (C) Seven independent experiments with distinct pDC donors (3–7 donors per condition) with n > 3 replicates per independent experiment. (D) Seven distinct pDC donor experiments (1–7 donors per condition) with n > 3 replicates per condition per experiment. The y axis represents IFN relative to baseline level generated after pDC + Huh coculture with 0.1 MOI DENV. Statistical significance was determined by 1-way ANOVA. *P < 0.05, ****P < 0.0001.
The cell-to-cell contact required for pDC sensing of DENV depends on the cell adhesion molecules ICAM-1 and αL-integrin, and prior data suggest that these cell adhesion molecules establish transient contact between pDCs and infected cells (60). If PAMP transfer occurs at these transient contact sites, long-term contacts are established, and TLR7-mediated IFN signaling occurs (60). To validate this, we treated infected Huh 7.5.1 with antibody blockade of ICAM-1 (Figure 1C) or αL-integrin (Figure 1D). We demonstrated that blocking either of these cell adhesion molecules results in the complete loss of IFN-α2a production, confirming a requirement for these adhesion molecules in pDC sensing (Figure 1, C and D).
Monoclonal anti-DENV antibodies against a subset of DENV epitopes enhance in vitro IFN production by pDCs. Given our previous study demonstrating that envelope-specific (E-specific) antibodies can alter innate immune signaling of HIV by pDCs (62), we hypothesized that preexisting antibodies in a DENV-exposed individual might alter the innate immune response to a subsequent heterotypic DENV or to subsequent ZIKV infection. To test this hypothesis, we assessed the effect of anti-DENV mAbs on pDC sensing of DENV in our in vitro model using a panel of previously characterized cross-reactive antibodies. We first used mAb DV87.1 (DVSF-3), previously defined as a DENV1 antibody specific for E domain III and cross-reactive for multiple DENV serotypes (64). To assess the effect of mAb DV87.1, DENV2-infected Huh 7.5.1 cells were incubated with anti-DENV DV87.1 for 1 hour and then cocultured with pDCs, with levels of IFN-α2a assessed after 24 hours. The addition of antibody after infection was designed to allow replication to occur in the absence of antibody to assess the effects of DENV-specific antibodies on sensing, rather than on infection. The results demonstrate that IFN production by pDCs is upregulated in the presence of anti-DENV mAb DV87.1 in a dose-dependent manner (Figure 2A). DV87.1 antibody did not trigger IFN-α2a production in a pDC/Huh 7.5.1 coculture system in the absence of virus, nor did it trigger IFN-α2a production when incubated with virus and then subsequently incubated with primary pDCs (Supplemental Figures 2 and 3).
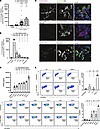 Figure 2
Figure 2Antibody-mediated IFN upregulation requires binding to a viral epitope on the surface of infected cells. Huh 7.5.1 cells were infected with DENV at 0.1 MOI for 48 hours. (A–C) After 48 hours, 0.1-1 μg/mL DV87.1 (A); 1 μg/mL DV87.1, 0.49 μg/mL C8, 0.29 μg/mL C10, 0.42 μg/mL 4G2, 20.4 μg/mL 2H2, or 1 μg/mL isotype control IgG (B); or 1 μg/mL 4G21, 1H10, 1B22, 2H21, 1E23, or DV87.1 (C) were added to infected Huh 7.5.1 cells and incubated for 1 hour. Primary human pDCs were added and cocultured for 24 hours. Supernatants were collected for IFN-α2a measurement. (D) Infected Huh 7.5.1 cells were treated with DV87.1 or 2H2 for surface binding of DENV epitopes; they were fixed and permeabilized after 24 hours and then stained intracellularly with 4G2, 2H2, or DV87.1. Scale bars: 10 μM. (E) Uninfected or DENV infected Huh 7.5.1 cells were surface stained with antibodies against DENV E or prM protein and stained with secondary anti–human AF647 or DY488. The left panels are representative flow plots for uninfected or infected Huh 7.5.1 cells. Right panels quantify flow data on left as percentage of DENV+ cells stained with each antibody. Significance was determined by comparing the binding of each antibody to infected cells over uninfected cells. Panels represent 17 (10–17 per condition) (A) and 4 (1–4 per condition) (B) independent experiments with unique pDC donors with n ≥ 3 per condition per experiment, C represents 1 independent experiment (conducted 4 times) with n ≥ 3 per condition, D is representative of 5 total experiments, and (E) represents 2 independent experiments with unique pDC donors for both the top and bottom panels. Statistical significance was determined by 1-way ANOVA. *P < 0.05, ***P < 0.001, ****P < 0.0001.
Having established that DV87.1 increases type I IFN production, we assessed a panel of broadly cross-reactive DENV-specific mAbs to assess epitope specificity of the effect. We used mAbs C8 and C10, human mAbs specific for DENV E epitope EDEI and previously characterized as highly DENV serotype cross-reactive and neutralizing (65). We also used the murine mAbs 2H2 and 4G2, specific for premembrane (prM) and E, respectively, and also cross-reactive for multiple DENV serotypes (66, 67). We assessed the ability of each mAb to promote pDC sensing of DENV as described in Figure 2A. C8 significantly increased IFN-α2a production, though not as robustly as DV87.1, while C10 showed a trend toward increased IFN-α2a production (P = 0.0867) (Figure 2B). In contrast, murine anti-E mAb 4G2 and anti-prM mAb 2H2 did not demonstrate a significant effect on IFN-α2a production (Figure 2B).
To determine whether the lack of effect of 2H2 on pDC sensing was due to this antibody’s specificity for prM and not E, we assessed an additional panel of human anti-prM antibodies (4G21, 1H10, 2H21, 1E23, and 1B22) previously characterized as being prM specific without cross-reactivity for E (characterized in Supplemental Table 1, adapted from ref. 68). The experiment was performed as described in Figure 2A, with a range of effects observed. prM-specific mAbs 4G21 and 1H10 did not significantly increase pDC IFN production relative to virus alone. However, 2H21, 1E23, and 1B22 significantly upregulated IFN generated by pDC sensing of DENV, although less robustly than DV87.1 (Figure 2C). These data demonstrate that mAbs targeting diverse DENV E and prM epitopes can enhance pDC sensing of DENV but that this is not a universal characteristic of all antibodies that target these proteins. Thus, it is clear that antigen specificity is not the only determinant of upregulated pDC sensing.
Modulation of IFN production by anti-DENV antibodies is dependent on the amount of Fab/epitope binding on the surface of infected cells. Although DENV virions are not thought to bud from infected cells (69), it has been shown that excess viral proteins stud the surface of DENV-infected cells (70–72). This suggests that DENV epitopes might be available at the surface of target cells and provide a target for anti-DENV antibodies. Given the range of enhancing effects of our mAbs targeting diverse E and prM epitopes, we hypothesized that the amount of mAb binding to the infected cell surface determines the capacity of the mAb to enhance pDC sensing.
We used 2 methods to test the capacity of our antibodies to bind cell surface antigen. The first method was surface immunofluorescence staining to assess whether the antibodies that enhanced IFN production also bind to the surface of DENV-infected Huh 7.5.1 cells. Surface-labeled cells were counterstained with a murine anti-E mAb (4G2) or a human anti-E mAb (DV87.1) to label intracellular DENV E protein and to identify successfully infected cells. Imaging of these stained cells revealed DENV E present on the surface of infected Huh 7.5.1 cells, labeled robustly by DV87.1 (Figure 2D). Staining was compared with the well-characterized surface stain wheat germ agglutinin to validate that the staining pattern observed was consistent with cell surface binding (Supplemental Figure 4). DV87.1, which is highly effective at upregulating IFN production, bound specifically to the surface of infected, but not uninfected, cells. Two additional mAb capable of enhancing pDC sensing of DENV, C8 and C10, bound the surface of infected cells robustly (Supplemental Figure 5). In contrast, the mAb 2H2, which did not enhance pDC sensing, was unable to bind the surface of infected cells despite being able to bind intracellular viral protein (Figure 2D). Collectively, these data support the hypothesis that enhanced pDC sensing of virally infected cells requires binding of an epitope present on the infected cell’s surface.
We used flow cytometric analysis of infected Huh 7.5.1 cells to detect surface DENV E and prM epitopes as a second method to verify these findings. DENV-infected cells were stained and analyzed for expression of prM and E on the cell surface and compared with uninfected controls; data are quantified as percentage of DENV+ cells for all tested mAbs (gating strategy as demonstrated Supplemental Figure 6). Surface staining with DV87.1, C8, and C10 confirmed that E epitopes were accessible on the surface of infected cells (Figure 2E). The degree to which these antibodies bind the surface of infected cells correlates with their ability to boost type I IFN production — namely, C8 and C10 bound much less robustly and also generated a less robust increase in IFN. Surface staining with 2H21, 1E23, and 1B22 confirmed that some prM epitopes were also accessible on the surface of infected cells. However, the prM-specific mAbs that did not significantly upregulate pDC sensing, 4G21 and 1H10, did not bind infected cells (Figure 2E), either because the epitopes were not expressed on the cell surface or the mAb avidity was too low to bind these epitopes on the cell surface. The dramatic differences in binding among anti-E and anti-prM antibodies suggest that epitope accessibility or antibody affinity for surface-expressed epitopes differ. Importantly, the degree to which a given antibody binds to infected cells as quantified by flow cytometry correlates with their IFN-promoting activity (Figure 2C) for the vast majority of antibodies in our panel, suggesting that upregulation of pDC-mediated IFN activity is, at least in part, defined by the ability of the antibody to bind to infected cells. These data support our hypothesis that antibody binding to viral epitopes accessible on the surface of infected cells is necessary to increase IFN production, with a direct correlation between the degree of antibody binding to the infected cell surface and the level of increased pDC IFN production.
We then established a hypothesis that antibodies can upregulate pDC IFN production in response to DENV infection by enhancing cell-to-cell contact between the sensing pDC and the DENV-infected cell, tethering the pDC specifically to an infected cell, as opposed to the nonspecific cell-to-cell interaction afforded by ICAM-1 engagement of αL-integrin.
Anti-DENV mAb facilitates the interaction of pDCs with infected cells. Our data, thus far, suggest that the Fab/epitope interaction on the surface of an infected cell is required for increased production of type I IFN by pDCs. To test our hypothesis that DENV antibodies that bind the surface of infected cells would lessen the dependence of pDC on adhesion molecules to sense infection, we reassessed the requirement for ICAM-1 and αL-integrin in the presence of these mAb (Figure 3A). Specifically, we treated DENV-infected Huh 7.5.1 cells with anti–αL-integrin antibody, followed by DV87.1 mAb. As in Figure 1B, treatment with anti–αL-integrin antibody in the absence of mAb led to a complete loss of IFN-α2a production by pDCs cocultured with infected Huh 7.5.1. However, the addition of the DV87.1 mAb rescued IFN-α2a production in a dose-dependent manner (Figure 3B). The same effect was observed when Huh 7.5.1 were cultured with anti–ICAM-1 and then treated with DV87.1 (Figure 3C). These data suggest that DENV-specific mAbs enhance pDC sensing of DENV by altering interactions between pDCs and infected cells, thereby reducing dependence on nonspecific adhesion molecules, potentially through higher affinity or prolonged cell-to-cell interactions.
 Figure 3
Figure 3Anti-DENV antibodies bypass the requirement for cell adhesion molecules in pDC sensing of virus-infected cells. (A) Schematic representation of experimental design. As established in Figure 1, blockade of ICAM-1 or αL-integrin abrogates IFN production by sensing pDCs. (B and C) Evaluation of the role of ICAM-1 or αL-integrin in the presence of anti-DENV antibody (represented in purple). Infected Huh 7.5.1 cells were incubated for 1 hour with anti-αL integrin at 0.1 μg/mL (B) or anti–ICAM-1 antibody at 5 μg/mL or 2 μg/mL (C) with or without DV87.1 at 0.1 μg/mL or 1 μg/mL. pDCs were added and cocultured for 24 hours. Supernatants were collected and assessed for IFN-α2a; B represents 1 pDC donor experiment with n ≥ 3 per condition, and C represents 11 pDC donor experiments (1–11 per condition) with n ≥ 3 per condition. Statistical significance was determined by 1-way ANOVA. *P < 0.05, **P < 0.01, ****P < 0.0001.
Modulation of IFN production by anti-DENV antibodies requires Fc/FcR interactions. Consistent with our proposed model, we hypothesized that Fc engagement of FcR on the sensing pDC is required for upregulated IFN production. Our previous research demonstrated that HIV mAbs enhance pDC sensing and IFN production in an Fab- and FcγR2a-dependent manner (62). We used 3 different methods to determine whether binding of the Fc portion of antibody to Fc-γ receptor (FcγR) is necessary for increased pDC sensing of DENV. First, we used a variant of the mAb DV87.1 with 2 leucine-to-alanine mutations that abrogate binding to FcγRs (LALA DV87.1) (64). In contrast to the WT DV87.1, the LALA DV87.1 mAb did not enhance IFN production when incubated with infected Huh 7.5.1 cells prior to coculture with pDCs (Figure 4A). Next, we used FcBlock to compete with the Fc portion of DV87.1 mAb for binding to all FcγRs on the pDC surface. When pDCs were preincubated with FcBlock, DV87.1 mAb failed to enhance IFN production (Figure 4B). Finally, we used a specific anti-FcγR2a (CD32a) blocking antibody to determine whether this effect was specifically dependent on this FcγR (62). When pDCs were preincubated with anti-FcγR2a, DV87.1 mAb no longer enhanced IFN-α2a production (Figure 4B). Taken together, these data support that Fc binding to FcγR, specifically FcγR2a, is required for antibody-mediated upregulation of type I IFN production by pDCs.
 Figure 4
Figure 4Antibody-mediated upregulation of pDC sensing and IFN production requires Fc engagement of FcR2a on pDCs. Huh 7.5.1 cells were infected with DENV at 0.1 MOI for 48 hours. (A) Infected Huh 7.5.1 were treated with DV87.1 or LALA DV87.1 antibody for 1 hour, followed by pDC coculture for 24 hours. (B) Infected hepatoma cells were cultured with DV87.1 for 1 hour, while pDCs were treated with FcBlock or anti-FcγR2a antibody for 1 hour. pDCs were then cocultured with Huh 7.5.1 for 24 hours. (C and D) Huh 7.5.1 were incubated with anti–ICAM-1 antibody at 2 μg/mL (C) or anti–αL integrin antibody at 0.1 μg/mL with or without DV87.1 (0.1 μg/mL or 1 μg/mL) or LALA DV87.1 (0.1 μg/mL or 1 μg/mL) (D). Afterward, they were cocultured with primary human pDCs for 24 hours. (E) Huh 7.5.1 cells were treated with anti–ICAM-1 blocking antibody at 2 μg/mL with 0.1 μg/mL or 1 μg/mL of DV87.1, while pDCs were treated with anti-FcγR2a blocking antibody at 10 μg/mL. (F) Infected Huh 7.5.1 cells were incubated with anti–ICAM-1 antibody at 2 μg/mL with 1 μg/mL of DV87.1 and increasing amounts of LALA DV87.1 (0.1 μg/mL or 10 μg/mL) for 1 hour; they were then cocultured with pDCs. All supernatants were collected and assessed for IFN-α2a by MSD analysis. Each panel represents 15 (A) and 3 (2–3 per condition) (B) independent experiments with unique pDC donors, with n ≥ 3 per condition per experiment. (C, E, and F) At least 2 independent experiments are shown, with unique pDC donors, and n ≥ 3 per condition per experiment. (D) A single pDC donor experiment is shown, with n ≥ 3 per condition. Statistical significance was determined by 1-way ANOVA. *P < 0.05, **P < 0.01, ****P < 0.0001.
To determine if bypassing cell adhesion molecules by antibody requires the engagement of FcγR2a on the pDC, as well as Fab binding to the infected cell, we assessed the role of FcγR in rescuing IFN production as in Figure 4, A and B. In contrast to DV87.1 mAb, the addition of LALA DV87.1 mAb failed to rescue IFN production in the presence of ICAM or αL-integrin blockade, suggesting that rescue is dependent on Fc/FcγR binding (Figure 4, C and D). To further test this requirement for FcγR engagement, we blocked pDC FcRs with anti-FcγR2a while treating infected Huh 7.5.1 with anti-ICAM antibody and DV87.1. In the presence of anti-FcγR2a, DV87.1 was incapable of rescuing IFN production, further validating that FcγR2a binding is critical to rescuing IFN production in the absence of adhesion molecule engagement (Figure 4E). Finally, we performed a competition experiment in which we treated infected Huh 7.5.1 with anti-ICAM antibody, followed by treatment with DV87.1 and increasing concentrations of LALA DV87.1. LALA DV87.1 mAb competes for E binding on the surface of infected hepatoma cells in a dose-dependent fashion but is incapable of binding pDC FcRs, including FcγR2a. The DV87.1 mAb binds FcR but is outcompeted for Fab binding in the presence of high amounts of LALA DV87.1. With increasing amounts of LALA DV87.1 added, the capacity for WT DV87.1 to rescue IFN production in the presence of ICAM blockade was abrogated (Figure 4F). The results of this competition experiment suggest that bridging the pDC to the infected cell via the same antibody molecule is necessary — not simply that Fab binding to infected cells and Fc binding to pDCs in isolation triggers increased IFN production. The Fab portions of these antibodies bind E protein epitopes accessible on the surface of infected cells with the Fc portion tethering the pDCs to these target cells and providing pDC specificity for infected target cells. This contrasts with a naive host environment in which random, nonspecific cell interactions must result in sufficient contact to trigger sensing. Anti-ICAM antibodies used extensively in our experiments do not mediate an increased IFN response to infected cells, as they are murine in origin and, thus, are not expected to bind human pDC FcR. Furthermore, the binding of anti-ICAM is not specific for infected cells.
Anti-DENV antibodies cross-reactive for ZIKV facilitate pDC sensing of ZIKV. The impact of anti-DENV antibodies in ZIKV immunity remains an open question. To address this, we assessed whether DENV-specific mAb altered type I IFN production in response to ZIKV-infected cells. Many mAbs binding to the EI or EII domains of DENV E or the E dimer epitope (EDE) regions of DENV are cross-reactive with ZIKV, providing the possibility for these antibodies to increase sensing of ZIKV. However, some mAbs directed to the less homologous EIII domain are not cross-reactive with ZIKV (14, 17). We assessed whether a subset of our panel of anti-DENV mAbs that boost the pDC response to DENV also augment pDC sensing of ZIKV-infected cells. C8 and C10 mAbs, both specific for the EDE1 region of DENV E and previously characterized as cross-reactive with ZIKV (14), increased IFN-α2a production when added to a coculture of pDCs and ZIKV Nicaragua– or ZIKV São José do Rio Preto–infected (SJRP-infected) Huh 7.5.1 cells (Figure 5, A and B). However, when the non–cross reactive mAb DV87.1 (an EIII-specific mAb) or 2H2 (an anti–prM DENV mAb) were added into the coculture system, neither upregulated IFN-α2a production in response to ZIKV (Figure 5, A and B). We then assessed the panel of mAbs for binding to the surface of ZIKV-infected Huh 7.5.1 cells, as tested previously with DENV-infected cells. Consistent with what we had observed for DENV, all antibodies that enhanced type I IFN production in the pDC coculture system (with C10 shown as a representative example here) bound the surface of ZIKV-infected cells, while those that did not increase IFN production (DV87.1, for example) did not bind the surface of ZIKV-infected cells (Figure 5C and Supplemental Figure 7). While DV87.1 bound the surface of DENV-infected cells and was highly upregulating of pDC-mediated DENV sensing (Figure 2A), it did not bind the surface of ZIKV-infected cells and did not boost IFN production in our ZIKV coculture system (Figure 5C), consistent with its binding to the EIII epitope, an epitope less conserved between DENV and ZIKV. Unlike DENV, ZIKV-infected Huh cells were not highly stimulating of pDC type I IFN in our assays. Thus, there is no baseline type I IFN production to rescue after cell adhesion molecule blockade. However, type I IFN upregulation in the presence of ZIKV Nicaragua or SJRP can be blocked in a dose-dependent fashion by anti-ICAM antibody when also cultured in the presence of C8 or C10 mAbs (Figure 5, D and E, and Supplemental Figure 8). These data are consistent with our model that antibody binding of epitopes expressed on the infected cell surface tether infected cells to pDC and increase IFN production from baseline. These data suggest that this phenomenon is not restricted to innate sensing of DENV and that antibodies generated in response to DENV may modulate the innate immune response to ZIKV. Notably, significant differences were not observed in the pDC response generated in the presence of antibodies between ZIKV strains.
 Figure 5
Figure 5Cross-reactive anti-DENV mAbs increase IFN production in pDC exposed to ZIKV. (A and B) Huh 7.5.1 cells were plated and infected with 0.1–1 MOI of ZIKV Nicaragua/2015 (A) or ZIKV SJRP/2015 (B) for 48 hours. Infected Huh 7.5.1 cells were replated and preincubated with anti-DENV mAbs C8, C10, 2H2, and DV87.1 for 1 hour. Primary human pDCs were added and cocultured with infected and antibody-treated Huh 7.5.1 cells for 24 hours, after which supernatants were assessed for IFN-α2a by MSD analysis. (C) Huh 7.5.1 cells were infected with ZIKV and then replated for 24 hours, at which point they were stained with DV87.1 and C10 mAbs for 1 hours at 4°C. Following surface Ab staining, the cells were fixed, permeabilized, and stained for intracellular DENV E with mouse mAb 4G2. Representative images for 4 conducted experiments are displayed (ZIKV SJRP used in displayed images). Scale bars: 10 μM. (D and E) Huh 7.5.1 cells were plated and infected with 0.1 MOI of ZIKV Nicaragua/2015 (D) or ZIKV SJRP/2015 (E) for 48 hours. Infected cells were replated and preincubated for 1 hour with the anti–ICAM-1 antibody at 0.1–5 μg/mL and C8 (D) or C10 (E) mAbs. After preincubation, primary human pDCs were isolated and cocultured with infected and antibody-treated Huh 7.5.1 cells for 24 hours, after which supernatants were assessed for IFN-α2a by MSD analysis. Each figure panel represents at least 2 independent experiments, with unique pDC donors, with n ≥ 3 per condition per experiment. Statistical significance was determined by 1-way ANOVA. ****P < 0.0001.
Polyclonal IgG isolated from individuals with immunity to DENV can alter pDC sensing of DENV and ZIKV. Having demonstrated that anti-DENV mAbs can alter the pDC response to both DENV and ZIKV, we next hypothesized that individuals exposed to DENV of 1 serotype might generate cross-reactive antibodies that can alter the type I IFN response to a second serotype of DENV or to ZIKV. To test this hypothesis across multiple DENV serotypes, we isolated polyclonal IgG (pIgG) from human participants enrolled in a randomized placebo-controlled double-blinded DENV challenge study (NCT02433652; clinicaltrials.gov) and assessed the effect of this pIgG on pDC sensing of DENV. Twenty-four individuals who were naive to DENV were randomized to receive either placebo (6 individuals) or vaccination (18 individuals) with a live attenuated admixture containing DENV1, DENV3, and DENV4 viruses. Twenty-one individuals (6 placebo recipients; 15 vaccine recipients) were then challenged with DENV2. Vaccinees were expected to generate a broad humoral response to DENV1, DENV3, and DENV4, and a subset would develop cross-reactive pIgG that binds DENV2. Given that plaque reduction neutralization titer 50% (PRNT50) antibody titers against DENV1–4 were highest on average at day 28 after vaccination, we isolated pIgG from the serum of vaccine and placebo recipients on day 28 and assessed their effect on our in vitro pDC/DENV-infected Huh 7.5.1 model. Total pIgG samples were isolated from 2 placebo recipients (placebo [P] participants P1 and P2), 6 vaccinees who were well protected after challenge (no detectable viremia by culture, vaccinees protected [VP] participants VP1–6), and 3 vaccinees who were not protected (detectable viremia, rash after challenge, vaccinees viremic [VV] participants VV1–3) and were assessed. Of these 11 individuals, pIgG from subject VP3, a well-protected vaccine recipient, strongly boosted type I IFN production in our in vitro assay, while pIgG from placebo recipients (participants P1 and P2) and from participants who were not well protected against challenge (VV1–3) did not (Figure 6A). However, pIgG from a subset of the participants protected from challenge also failed to enhance IFN production, demonstrating that antibodies induced by vaccination and challenge are not uniformly able to increase IFN production (Figure 6A). When these pIgG were assessed for their ability to restore IFN production in the presence of cell adhesion molecule blockade, IgG from subject VP3 was the only pIgG capable of rescuing pDC sensing of DENV-infected cells (Figure 6B). This is consistent with the pattern established with mAbs, in which antibodies that upregulated IFN activity were capable of rescuing IFN production when ICAM/integrin interactions were blocked (Figure 6B).
 Figure 6
Figure 6Polyclonal IgG from DENV-immune individuals can enhance IFN activity of pDC exposed to DENV or ZIKV. (A) Huh 7.5.1 cells were infected with 0.1 MOI of DEN2-NGC for 48 hours. Polyclonal IgG isolated from DENV-vaccinated (participants VV1–3, VP1–6) or DENV-naive (participants P1, P2) participants 28 days after vaccination, with live attenuated admixture of DENV1, DENV3, and DENV4 or placebo added to culture after 48 hours. VV represents vaccinated and viremic participants, VP represents vaccinated, and protected participants, and P represents placebo-treated participants. After 1 hour of preincubation with polyclonal IgG, primary human pDCs were cocultured for 24 hours. IFN-α2a was assessed in the supernatant. The figure represents 2 independent pDC donor experiments, with n ≥ 2 per condition. (B) Infected Huh 7.5.1 cells were preincubated with the anti–ICAM-1 antibody at 2 μg/mL for 1 hour. Polyclonal IgG isolated from DENV-vaccinated (participants VV1, VP1–6) were added for 1 hour. Primary human pDCs were isolated and cocultured for 24 hours. IFN-α2a was assessed in supernatants. (C) Huh 7.5.1 cells were infected with ZIKV SJRP or ZIKV Nicaragua for 48 hours. They were then cultured with polyclonal IgG isolated from DENV-immune participants (participants VV1, VP1–4) for 1 hour and then cocultured with pDCs for 24 hours. IFN-α2a was measured in the supernatant. (B and C) A single pDC donor experiment is shown, with n ≥ 3 per condition. (D) Infected Huh 7.5.1 cells were treated with polyclonal IgG from VP3, P1, and VP4 to assess binding of polyclonal IgG to the surface of infected cells. Cells were fixed, permeabilized, and stained intracellularly with 4G2 murine anti-DENV E antibody. Representative images for 4 total experiments are displayed. Scale bars: 10 μM. Statistical significance was determined by 1-way ANOVA. ****P < 0.0001.
We next assessed the pIgG of these same individuals in our model of ZIKV sensing to assess whether the effects of monoclonal anti-DENV antibodies on ZIKV sensing might be replicated by polyclonal antibodies (pAbs) generated in individuals exposed to DENV. When we cocultured pIgG from the same individuals described above, we observed the same patterns of altered IFN-α2a production. pIgG from subject VP3 strongly upregulated IFN-α2a production in our assay in response to ZIKV, while IgG from placebo recipients and other vaccine recipients did not significantly alter IFN-α2a production (Figure 6C).
To validate whether the modulation of type I IFN by pIgG is consistent with the mechanism demonstrated with mAbs, we assessed the isolated pIgG for binding to the surface of infected cells via immunofluorescence as performed with mAb against DENV. As predicted, pIgG from subject VP3 robustly bound the surface of DENV-infected cells, while pIgG isolated from participants that did not increase IFN production did not bind the surface of these cells (Figure 6D). These data show that, as with mAb, there is a heterogeneity in the ability of pIgG generated in DENV vaccination to increase IFN in response to subsequent DENV or ZIKV infection.
-
Discussion
The type I IFN pathway is critical in establishing the early antiviral immune response to DENV and ZIKV by directly restricting viral replication and shaping the downstream adaptive immune response. Our data suggest that anti-DENV antibodies that bind specifically to epitopes accessible on the surface of DENV- or ZIKV-infected cells increase the type I IFN response by pDCs independently of neutralization potential. Antibody-mediated upregulation of type I IFN production by pDCs requires Fab binding to an epitope present on infected cells and Fc-FcγR2a engagement on the pDC. Our work suggests that virus-specific antibody can function instead of or in concert with the naturally occurring ICAM-1/αL-integrin interactions to bridge the pDC to the infected cell, leading to more specific cell-to-cell contacts and greater IFN production (Figure 7). We also show that the presence of cross-reactive anti-DENV antibodies increased type I IFN production when pDCs interacted with ZIKV-infected cells, offering information about cross-reactive immunity between DENV and ZIKV. Finally, as demonstrated with mAbs, we have shown that anti-DENV pIgG from an individual given a live attenuated DENV1, DENV3, and DENV4 vaccine increased the production of type I IFN by pDCs in response to serotype 2 DENV and ZIKV infection. In summary, these data suggest a role for nonneutralizing antibodies to offer protection via modulation of the innate immune response to DENV and ZIKV.
 Figure 7
Figure 7A proposed model for antibody-mediated specificity in pDC sensing of DENV and subsequently enhanced IFN production. In the absence of anti-DENV antibodies, random interactions (represented on the left side of the schematic) result in pDCs interacting with infected and uninfected cells randomly, forming transient contacts via ICAM-1/integrin interactions. When a pDC randomly encounters an infected cell, PAMP transfer occurs, and TLR7 signaling and IFN production ensue. When the anti-DENV antibody is present (represented on the right side of the schematic), antibody-directed contact with DENV- or ZIKV-infected cells occurs. This mechanism works in concert with ICAM/integrin interactions to bind pDCs specifically to the infected cell, possibly increasing the number of pDC interactions with infected cells, the duration, or the avidity of pDC interactions with infected cells.
We propose that upregulation of type I IFN production is not the sole, but one of several, mechanisms of protection provided by cross-reactive antibodies. DENV1-, DENV3-, and DENV4-vaccinated individuals were protected from severe infection with heterotypic DENV2. One of the individuals who was well protected without detectable viremia by culture following vaccination generated pIgG that mediated increased type I IFN production and bound the surface of infected cells, as measured by immunofluorescence, suggesting that the described phenomenon may have contributed to viral control. However, pIgG isolated from other protected individuals failed to alter IFN production and did not bind the surface of infected cells, indicating that this is not the only, but likely one of many, mechanisms of protection that may include neutralizing antibodies and T cell responses resulting from vaccination. Our data highlight that the polyclonal response to DENV infection is composed of antibodies with diverse features and that the resulting population of antibodies may shape the individual’s clinical outcome through neutralizing and nonneutralizing means.
Our model demonstrates dependence on the availability of viral epitopes on the surface of both DENV- and ZIKV-infected cells for IFN enhancement. The accessibility of DENV epitopes at the surface of infected cells could be the result of viral protein accumulation at the plasma membrane or of binding of intact virions to the cell membrane, either on entry into a target cell or on exocytosis from an infected cell. Our data do not distinguish between these possibilities; however, the existing literature and our current data demonstrate that viral epitopes are present on the infected cell surface (70–72). Furthermore, our data that pDCs do not produce IFN when cultured with virus preincubated with antibody demonstrate that IFN enhancement by anti-DENV antibodies is not due to enhanced pDC internalization of free virus. This supports the proposed model and the dependence on interactions between pDCs and infected cells expressing viral epitopes on the cell surface.
Our data extend the existing literature on the nonneutralizing effects of antibody responses to DENV and ZIKV. Monocytic cells, a secondary producer of type I IFN in DENV infection, have been the focus of previous studies and show reduced type I IFN production in the presence of preexisting cross-reactive anti-DENV antibodies, thus worsening measured viral infection (73–75). However, the primary type I IFN–producing cell in many viral infections is the pDC (51–53), making the pDC critical in our understanding of the type I IFN response in secondary DENV or ZIKV infection. Our data are consistent with the limited literature that exists on pDC response to DENV, as well as the data on the in vivo IFN response after exposure (57–61). Specifically, studies demonstrating that people with severe DENV have decreased peripheral IFN and a suppressed pDC response support a mechanism of protection by which preexisting antibodies upregulate IFN production (57).
Our data demonstrate, for the first time to our knowledge, the effect of cross-reactive antibodies on the pDC viral sensing pathway following DENV or ZIKV exposure and add to existing literature establishing the concept of the interferogenic synapse in pDC sensing of DENV and ZIKV (59, 60). This antibody effector mechanism may be relevant in protection against a variety of viruses, as pDCs generate large quantities of type I IFNs when they interact with liver cells infected with DENV, ZIKV, and other important pathogens (59, 61). The released IFNs directly function to restrict viral replication (29, 30, 61). We established that a specific subset of cross-reactive antibodies increased IFN production, leading to more robust innate immune protection relative to primary infection in a mechanism that relies on contact between innate sensing and infected cells. However, our data do not distinguish whether the antibody-mediated increase in IFN production results from increasing the number or the duration of pDC/infected cell contacts.
Our work defines a mechanism independent of neutralization by which cross-reactive antibodies present at the time of secondary DENV or ZIKV infection might be protective. Our in vitro model specifically utilizes a system in which infection and replication occur for 48 hours prior to the addition of antibodies, bypassing the infection neutralization effect of these antibodies. Importantly, the ability of an antibody to neutralize DENV or ZIKV infection did not segregate completely with IFN upregulation, and notably, not all neutralizing antibodies upregulate IFN production. It is well known that nonneutralizing antibodies can enhance disease severity, but the vast majority of secondary DENV infections are not severe. Antibodies that increase IFN production may mitigate the risk of worsened clinical outcomes in a subset of individuals with antibodies to DENV. These data further highlight the complexity of and potential for diverse effects of antibodies in clinical outcomes.
We have demonstrated that the IFN innate immune signaling cascade can be upregulated in DENV and ZIKV infections in vitro by the presence of preexisting cross-reactive anti-DENV antibodies, suggesting a possible mechanism for enhanced protection against these viruses following primary infection. This work advances our understanding of the role of nonneutralizing antibodies in viral infection, flavivirus, and otherwise — particularly in understanding the interplay between antibody and innate immune responses. Furthermore, it highlights the multifaceted role of antibodies in clinical outcomes.
-
Methods
Supplemental Methods are available online with this article.
Expansion of DENV and ZIKV cell culture strains. DENV2-NGC, ZIKV Nicaragua/2015, and ZIKV SJRP/2015 were obtained from NIAID. The viruses were amplified as has been previously described (76). Vero cells were plated and infected with 0.01 MOI of DENV2-NGC or 0.001 MOI of ZIKV SJRP/2015 or ZIKV Nicaragua/2015. Infected cells were cultured for 5–7 days. Vero culture supernatant was collected and aliquoted. PFU/mL was determined by a modified plaque assay and immunostaining. Briefly, Vero cells were plated in 24-well plates and cultured to 90% confluency. Virus was plated in duplicate in serum-free OPTIMEM at 6 serial 10-fold dilutions and incubated for 1 hour. An overlay of 1% methylcellulose in OPTIMEM was applied to cultures. Plates were incubated for 4–5 days. Titration plates were fixed with methanol and stained with anti-DENV antibodies (2H2 and 4G2, supplied by NIAID), followed by a secondary goat anti–mouse HRP–conjugated antibody (VWR, 074-1806). TrueBlue KPL substrate (VWR, 50-78-02) was used to develop plaques.
DENV mAbs. The following antibodies were obtained from NIAID, originally produced by LakePharma: 4G2, C8, and C10 (65, 66, 77, 78). Stephen Whitehead also provided the 2H2 antibody, originally purchased from the ATCC as hybridoma D3-2H2-9-21 (ATCC, HB-114). The following antibodies were obtained from Wistar Institute: pDV87.1, pDV87.1 LALA (64). pDV87.1 LALA is identical to pDV87.1 except for 2 leucine-to-alanine mutations (L234A, L235A) in the Fc portion of the antibody, which abrogate its binding to FcR (79). The following antibodies were obtained from Aravinda de Silva (University of North Carolina at Chapel Hill, North Carolina, USA): 4G21, 1H10, 2H21, 1E23, and 1B22 (68).
pIgG from DENV challenge cohort participants. Serum samples were generously shared from the NIAID Trivalent Vaccination Cohort Study and were obtained from the 2 sites involved in the cohort: Johns Hopkins University and the University of Vermont (Clinicaltrials.gov NCT02433652). The study is a challenge study designed to assess the efficacy of a trivalent live attenuated DENV vaccine formulation containing DENV1, DENV3, and DENV4 vaccine components and was sponsored by NIAID contract no. HHSN272200900010C. Participants were vaccinated or received placebo on day 0 of the study. On day 180, participants were all challenged with the recombinant DENV2 challenge virus rDEN2Δ30. Of the 25 participants enrolled in the cohort, 20 completed the course of the challenge study. We obtained day 28 postvaccination/placebo serum for 16 individuals (14 vaccinees and 2 placebo recipients). pAb was isolated using the Pierce Protein A IgG isolation columns (Thermo Fisher Scientific, 44667) per the manufacturer’s instructions. Each 1 mL elution fraction was neutralized using 100 μL of binding buffer. Fractions containing IgG as determined by nanodrop and SDS-PAGE were concentrated on a 50 kDa Amicon Ultra 5 mL filter (MilliporeSigma, UFC805024) and quantified using Nanodrop Protein A280 measurement.
Cell lines and primary pDC isolation. Huh 7.5.1 hepatoma cells were originally obtained from Dr. Charles Rice (Rockefeller Institute, New York, New York, USA) and were cultured in DMEM with 10% FBS and 1% nonessential amino acids. For primary human cell culture and pDC isolation, freshly collected deidentified human blood leukopaks (LPs) were obtained from the Anne Arundel Medical Blood Donor Center (Parole, Maryland, USA). PBMCs were isolated using Ficoll-Hypaque gradient centrifugation at 400g for 30 minutes at room temperature without brake. PBMCs were then subjected to magnetic separation, and pDCs were isolated by negative selection, per the manufacturer’s protocol (Miltenyi Biotec, 130-097-415). pDCs were collected and cultured in RPMI 1640 media supplemented with 10% FBS, 1% L-glutamine, 1% nonessential amino acids, 1% sodium pyruvate, and 1% HEPES buffer.
Coculture pDC experiments. On day 0, Huh 7.5.1 were plated at approximately 6 × 105 cells/well in a 6-well cell culture plate. Huh 7.5.1 cells carry a point mutation in the gene encoding RIG-I, which reduces the host innate immune response to viral RNA and renders cells more permissive to viral infection (80, 81). On day 1, Huh 7.5.1 were confluent at approximately 1.5 × 106 cells/well. Huh 7.5.1 were infected on day 1 with 0.1–1 MOI of DENV2-NGC or ZIKV or were left in media alone and cultured for 48 hours. On day 3, Huh 7.5.1 cells were lifted and replated at 100,000 cells/well in a 96-well U-bottom cell culture plate. After allowing cells to readhere, infected or uninfected Huh 7.5.1 were treated with cell culture compounds or antibody for 1 hour. Primary human pDCs were isolated as described above. pDCs were plated at 20,000 cells/well in a 96-well U-bottom cell culture plate. As needed, pDCs were treated with any cell culture compounds or antibodies for 1 hour. Finally, pDCs were transferred to coculture with Huh 7.5.1 for 24 hours. After 24 hours, cell culture supernatants were collected for further analysis.
IFN-α measurements. Human IFN-α2a was quantified using the Human IFN-α2a Tissue Culture Kit from Meso Scale Discovery (MSD, K151ACB-4). Samples were assessed per the manufacturer’s protocol, using 25 μL of supernatant, diluted 1:10 in pDC tissue culture media (described above). Data were acquired on a MESO QuickPlex SQ 120 imager. Data were analyzed using Meso Discovery Workbench software.
Modulation of IFN by antibody. As described above, Huh 7.5.1 cells were replated on day 3 of the experiment. After adherence in 96-well cell culture plates, 100,000 mock-infected or infected Huh 7.5.1/well were incubated with mAbs at 0.1 μg/mL-10 μg/mL or pAbs at 0.001 μg/mL-1000 μg/mL for 1 hour. Concentrations of DENV anti-E mAbs were selected based on their enhancement activity in the K562 assay. Following antibody incubation with mock-infected Huh 7.5.1 cells or with DENV2-NGC– or ZIKV-infected Huh 7.5.1 cells, pDCs were then cocultured as described above for 24 hours, and IFN-α2a was measured in supernatant to assess the effect of antibody on IFN production.
Modulation of IFN by cell adhesion molecules. Mock-infected or DENV2-NGC–infected Huh 7.5.1 cells were replated on day 3. After adherence, cell adhesion molecules were blocked for 1 hour using anti–ICAM-1 (CD54) antibody (Thermo Fisher Scientific, 559047) or anti–αL-integrin (ITGAL) antibody (LifeSpan Biotechnologies, LS-C134275-100) as described previously (60). After 1 hour, pDCs were cocultured for 24 hours, and IFN-α2a was measured in the supernatant following coculture.
Assessing FcR requirement. Freshly isolated pDCs were plated at 20,000 cells/well in a 96-well U-bottom cell culture plate. Prior to coculture, they were preincubated with one of the following reagents to assess FcR usage: FcBlock (BD Biosciences, 564219) or anti-FcγR2a (CD32a) antibody (R&D Systems, AF1875). Following the blockade of FcRs, pDCs were cocultured with Huh 7.5.1, which had simultaneously been incubated with a monoclonal antibody. In addition to FcR blockade, LALA variant DV87.1 antibody (which contains intact Fab but Fc regions unable to bind FcR) was used to further assess Fc/FcR binding requirements.
Immunofluorescent imaging of DENV-infected cells. Huh 7.5.1 hepatoma cells were infected with 0.1 MOI of DENV2-NGC. After 48 hours of infection, cells were replated onto coverslips and then blocked with PBS supplemented with Ca2+ and Mg2+ with 1% BSA. Then, cells were cultured with the given mAb or pAb (human mAbs DV87.1, C8, and C10; murine mAbs 2H2 and 4G2; or pIgG) in blocking buffer for 1 hour at 4°C to prevent endocytosis and allow for surface staining. Cells were fixed with 4% PFA and permeabilized using TBS with 1% BSA, 0.2% milk, and 0.15% saponin. Intracellular DENV was then stained using a second, different anti-DENV mAb diluted in permeabilization buffer. If surface staining was completed using a human mAb, intracellular staining was done with a murine mAb and vice versa. Stained cells were washed and stained with goat anti–human Dylight 650 (Thermo Fisher Scientific, MA1-016-D650), goat anti–human IgG AF488 (Invitrogen, A-11013), donkey anti–mouse AF488 (Invitrogen, A21202), or goat anti–mouse IgG AF594 (Abcam, ab150116). When relevant, cells were incubated for 10 minutes at 37°C and 5% CO2 with 5 μg/mL wheat germ agglutinin-AF594 (Invitrogen, W11262) prior to primary antibody incubation or fixation/permeabilization. Cells were counterstained with DV87.1 and a goat anti–human IgG AF488 secondary antibody (Invitrogen, A-11013). Images were acquired using a LSM800 confocal (Zeiss) with gallium arsenide phosphide (GaAsP) detectors.
Flow cytometry. Uninfected or DENV NGC–infected Huh 7.5.1 cells were gently scraped and washed 1× in cold PBS. Cells were stained for viability with Live/Dead Fixable Aqua dye 1:200 (Thermo Fisher Scientific) + FcBlock 1:20 (BD Biosciences) for 20 minutes at 4°C in the dark. Cells were washed 1× with cold PBS. Cells were then surface stained with human anti–DENV E antibodies DV87.1 (1 μg/mL), C8 (0.49 μg/mL), C10 (0.29 μg/mL), or human anti-prM antibodies 4G21, 1H10, 2H21, 1E23, or 1B22 (1 μg/mL) in 100 μL staining buffer (cold PBS + 0.5% BSA) for 30minutes at 4°C in the dark. Cells were washed 2× with cold staining buffer. Primary antibody stained cells were then stained with secondary anti–human AF647 1:1000 (Southern Biotech, 2048-31) or secondary anti–human Dy488 1:1000 (Abcam, ab 97003) in 100 μL cold staining buffer for 20 minutes at 4°C in the dark. Finally, cells were fixed in 1% paraformaldehyde and run on a 5-laser BD Biosciences Fortessa flow cytometer or a 4-laser Cytek Aurora. Analysis was performed using Flowjo v10 software.
Statistics. One-way ANOVA was performed with GraphPad Prism 7.0 software to assess statistical significance. Sidak’s or Tukey’s test was used to correct for multiple comparisons. Differences between groups were considered significant when P < 0.05. Data represent mean ± SEM. Data for many experiments were normalized as a percentage of the average of replicates of a baseline condition for the experiment (most often virus alone) due to the marked donor-to-donor variability in pDC IFN production; normalization is noted in all figures and figure legends when applicable.
Study approval. The use of human participants occurred via the NIAID Trivalent Vaccination Cohort Study (Clinicaltrials.gov, NCT02433652). The study was sponsored by NIAID under contract no. HHSN272200900010C, was approved by WIRB and the University of Vermont IRB, and was conducted under an FDA investigational new drug application.
-
Author contributions
LKA designed experiments, conducted experiments, acquired data, analyzed data, and wrote the manuscript. KER conducted experiments, acquired data, contributed to writing the manuscript, and analyzed data. KC conducted experiments, acquired data, and analyzed data. GM conducted experiments and acquired data. WHA assisted in conducting experiments, specifically immunofluorescent imaging studies, and helped to design these experiments. WL provided many reagents and helped in designing experiments. KM and DBW provided critical reagents. SSW assisted in designing experiments and shared critical reagents. MAC designed experiments and analyzed data. APD designed research studies and experiments, analyzed data, provided critical reagents, and contributed to the manuscript. ALC designed research studies and experiments, analyzed data, and contributed to writing the manuscript. The listing order of co–first authors was determined by alphabetical order.
-
Acknowledgments
We would like to thank the laboratory of Aravinda de Silva for providing critical anti-prM antibodies and LaToya Roker at the Johns Hopkins School of Medicine Microscope Facility for assistance with confocal imaging. We would like to acknowledge the following sources of funding for their support in completing this work: R01AI108403 (AC), 1U19AI159822 (AC), K08AI102696 (MC), BMGF- Zika grant (DBW), and the Intramural Research Program of the NIH, NIAID. Funding for CIR300 came from NIAID contract HHSN2722009000 (AD).
Address correspondence to: Andrea L. Cox, 855 North Wolfe Street, Room 551, Baltimore, Maryland 21205, USA. Phone: 410.502.2715; Email: acox@jhmi.edu.
-
Footnotes
Conflict of interest: ALC serves as a consultant for Janssen. DBW receives grant/research support, serves as a consultant, and is a stock shareholder managed for Geneos; receives grant/research support and serves as a consultant for GeneOne; receives grant/research support, and is a board member, shareholder, and consultant for Inovio; serves as a consultant for AstraZeneca; and serves as a consultant for Sanofi. APD serves as a consultant for Merck and Co. ALC’s and APD’s conflicts are disclosed to and managed by the Johns Hopkins University Office of Policy Coordination. DBW’s conflicts are disclosed to and managed by the Wistar General Councils Office.
Copyright: © 2022, Aisenberg et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(12):e151782.https://doi.org/10.1172/jci.insight.151782.
-
References
- Vorou R. Zika virus, vectors, reservoirs, amplifying hosts, and their potential to spread worldwide: what we know and what we should investigate urgently. Int J Infect Dis. 2016;48:85–90.
- Wang E, et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74(7):3227–3234.
- Centers for Disease Control and Prevention. Dengue — Statistics and Maps. https://www.cdc.gov/dengue/epidemiology/index.html Accessed May 23, 2022.
- Centers for Disease Control and Prevention. Zika Virus. https://www.cdc.gov/zika/about/overview.html Accessed May 23, 2022.
- Sirohi D, Kuhn RJ. Zika virus structure, maturation, and receptors. J Infect Dis. 2017;216(suppl 10):S935–S944.View this article via: PubMed Google Scholar
- Kostyuchenko VA, et al. Structure of the thermally stable Zika virus. Nature. 2016;533(7603):425–428.
- Suwanmanee S, Luplertlop N. Dengue and Zika viruses: lessons learned from the similarities between these Aedes mosquito-vectored arboviruses. J Microbiol. 2017;55(2):81–89.
- Dowd KA, et al. Broadly neutralizing activity of Zika virus-immune sera identifies a single viral serotype. Cell Rep. 2016;16(6):1485–1491.
- Katzelnick LC, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358(6365):929–932.
- World Health Organization. Dengue and Severe Dengue. https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue Accessed May 23, 2022.
- Guzman MG, Harris E. Dengue. Lancet. 2015;385(9966):453–465.
- Zellweger RM, et al. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7(2):128–139.
- Williams KL, et al. Therapeutic efficacy of antibodies lacking FcγR against lethal dengue virus infection is due to neutralizing potency and blocking of enhancing antibodies. PLOS Pathog. 2013;9(2):e1003157.
- Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol. 2016;17(9):1102–1108.
- Gunawardana SA, Shaw RH. Cross-reactive dengue virus-derived monoclonal antibodies to Zika virus envelope protein: Panacea or Pandora’s box? BMC Infect Dis. 2018;18(1):641.
- Khandia R, et al. Modulation of dengue/Zika virus pathogenicity by antibody-dependent enhancement and strategies to protect against enhancement in Zika virus infection. Front Immunol. 2018;9:597.
- Priyamvada L, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A. 2016;113(28):7852–7857.
- Fagbami AH, et al. Cross-infection enhancement among African flaviviruses by immune mouse ascitic fluids. Cytobios. 1987;49(196):49–55.View this article via: PubMed Google Scholar
- Bardina SV, et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017;356(6334):175–180.
- Montoya M, et al. Longitudinal analysis of antibody cross-neutralization following Zika virus and dengue virus infection in Asia and the Americas. J Infect Dis. 2018;218(4):536–545.
- McCracken MK, et al. Impact of prior flavivirus immunity on Zika virus infection in rhesus macaques. PLoS Pathog. 2017;13(8):e1006487.
- Abbink P, et al. Therapeutic and protective efficacy of a dengue antibody against Zika infection in rhesus monkeys. Nat Med. 2018;24(6):721–723.
- Barba-Spaeth G, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536(7614):48–53.
- Swanstrom JA, et al. Dengue virus envelope dimer epitope monoclonal antibodies isolated from dengue patients are protective against Zika virus. mBio. 2016;7(4):e01123–16.View this article via: PubMed Google Scholar
- Pantoja P, et al. Zika virus pathogenesis in rhesus macaques is unaffected by pre-existing immunity to dengue virus. Nat Commun. 2017;8:15674.
- Terzian ACB, et al. Viral load and cytokine response profile does not support antibody-dependent enhancement in dengue-primed Zika virus-infected patients. Clin Infect Dis. 2017;65(8):1260–1265.
- Delgado FG, et al. Improved immune responses against Zika virus after sequential dengue and Zika virus infection in humans. Viruses. 2018;10(9):E480. View this article via: PubMed Google Scholar
- Rodriguez-Barraquer I, et al. Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science. 2019;363(6427):607–610.
- Diamond MS, et al. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74(11):4957–4966.
- Diamond MS, Harris E. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology. 2001;289(2):297–311.
- Jiang D, et al. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol. 2010;84(16):8332–8341.
- Goebel S, et al. A sensitive virus yield assay for evaluation of antivirals against Zika virus. J Virol Methods. 2016;238:13–20.
- Tsai CY, et al. Type I IFNs and IL-18 regulate the antiviral response of primary human γδ T cells against dendritic cells infected with dengue virus. J Immunol. 2015;194(8):3890–3900.
- Muñoz-Jordán JL, et al. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100(24):14333–14338.
- Jones M, et al. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol. 2005;79(9):5414–5420.
- Mazzon M, et al. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200(8):1261–1270.
- Rodriguez-Madoz JR, et al. Dengue virus inhibits the production of type I interferon in primary human dendritic cells. J Virol. 2010;84(9):4845–4850.
- Manokaran G, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350(6257):217–221.
- Grant A, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19(6):882–890.
- Hertzog J, et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur J Immunol. 2018;48(7):1120–1136.
- Lin S, et al. Zika virus NS5 protein antagonizes type I interferon production via blocking TBK1 activation. Virology. 2019;527:180–187.
- Zheng Y, et al. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. EMBO J. 2018;37(18):e99347. View this article via: PubMed Google Scholar
- Ngueyen TTN, et al. Zika virus proteins NS2A and NS4A are major antagonists that reduce IFN-β promoter activity induced by the MDA5/RIG-I signaling pathway. J Microbiol Biotechnol. 2019;29(10):1665–1674.
- Simmons CP, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195(8):1097–1107.
- Popper SJ, et al. Temporal dynamics of the transcriptional response to dengue virus infection in Nicaraguan children. PLoS Negl Trop Dis. 2012;6(12):e1966.
- Long HT, et al. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. J Infect Dis. 2009;199(4):537–546.
- John DV, et al. Biomarkers of severe dengue disease — a review. J Biomed Sci. 2015;22(1):83.
- Chen J, et al. Outcomes of congenital Zika disease depend on timing of infection and maternal-fetal interferon action. Cell Rep. 2017;21(6):1588–1599.
- Esser-Nobis K, et al. Comparative analysis of African and Asian lineage-derived Zika virus strains reveals differences in activation of and sensitivity to antiviral innate immunity. J Virol. 2019;93(13):e00640–19.View this article via: PubMed Google Scholar
- Chehimi J, et al. Dendritic cells and IFN-alpha-producing cells are two functionally distinct non-B, non-monocytic HLA-DR+ cell subsets in human peripheral blood. Immunology. 1989;68(4):488–490.View this article via: PubMed Google Scholar
- Fitzgerald-Bocarsly P. Human natural interferon-alpha producing cells. Pharmacol Ther. 1993;60(1):39–62.
- Sandberg K, et al. Flow cytometric analysis of natural interferon-alpha producing cells. Scand J Immunol. 1991;34(5):565–576.
- Rönnblomo L, et al. Properties of human natural interferon-producing cells stimulated by tumor cell lines. Eur J Immunol. 1983;13(6):471–476.
- Reizis B, et al. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol. 2011;29(1):163–183.
- Gandini M, et al. Dengue virus activates membrane TRAIL relocalization and IFN-α production by human plasmacytoid dendritic cells in vitro and in vivo. PLoS Negl Trop Dis. 2013;7(6):e2257.
- Sun P, et al. Functional characterization of ex vivo blood myeloid and plasmacytoid dendritic cells after infection with dengue virus. Virology. 2009;383(2):207–215.
- Pichyangkul S, et al. A blunted blood plasmacytoid dendritic cell response to an acute systemic viral infection is associated with increased disease severity. J Immunol. 2003;171(10):5571–5578.
- Lertjuthaporn S, et al. Identification of changes in dendritic cell subsets that correlate with disease severity in dengue infection. PLoS One. 2018;13(7):e0200564.
- Décembre E, et al. Sensing of immature particles produced by dengue virus infected cells induces an antiviral response by plasmacytoid dendritic cells. PLoS Pathog. 2014;10(10):e1004434.
- Assil S, et al. Plasmacytoid dendritic cells and infected cells form an interferogenic synapse required for antiviral responses. Cell Host Microbe. 2019;25(5):730–745.
- Webster B, et al. Plasmacytoid dendritic cells control dengue and Chikungunya virus infections via IRF7-regulated interferon responses. Elife. 2018;7:e34273.
- Veenhuis RT, et al. HIV-antibody complexes enhance production of type I interferon by plasmacytoid dendritic cells. J Clin Invest. 2017;127(12):4352–4364.
- Serrano-Collazo C, et al. Effective control of early Zika virus replication by dengue immunity is associated to the length of time between the 2 infections but not mediated by antibodies. PLoS Negl Trop Dis. 2020;14(5):e0008285.
- Flingai S, et al. Protection against dengue disease by synthetic nucleic acid antibody prophylaxis/immunotherapy. Sci Rep. 2015;5:12616.
- Rouvinski A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520(7545):109–113.
- Kaufman BM, et al. Monoclonal antibodies for dengue virus prM glycoprotein protect mice against lethal dengue infection. Am J Trop Med Hyg. 1989;41(5):576–580.
- Dejnirattisai W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328(5979):745–748.
- Smith SA, et al. Dengue virus prM-specific human monoclonal antibodies with virus replication-enhancing properties recognize a single immunodominant antigenic site. J Virol. 2016;90(2):780–789.
- Rodenhuis-Zybert IA, et al. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci. 2010;67(16):2773–2786.
- Hase T, et al. An electron and immunoelectron microscopic study of dengue-2 virus infection of cultured mosquito cells: maturation events. Arch Virol. 1987;92(3–4):273–291.View this article via: PubMed Google Scholar
- Mackenzie JM, et al. Improved membrane preservation of flavivirus-infected cells with cryosectioning. J Virol Methods. 1996;56(1):67–75.
- Ng ML, Corner LC. Detection of some dengue-2 virus antigens in infected cells using immuno-microscopy. Arch Virol. 1989;104(3–4):197–208.View this article via: PubMed Google Scholar
- Yong YK, et al. Aberrant monocyte responses predict and characterize dengue virus infection in individuals with severe disease. J Transl Med. 2017;15(1):121.
- Flipse J, et al. Molecular mechanisms involved in antibody-dependent enhancement of dengue virus infection in humans. Traffic. 2013;14(1):25–35.
- Ubol S, Halstead SB. How innate immune mechanisms contribute to antibody-enhanced viral infections. Clin Vaccine Immunol. 2010;17(12):1829–1835.
- Durbin AP, et al. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3’-untranslated region. Am J Trop Med Hyg. 2001;65(5):405–413.
- Falconar AK. Identification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteins. Arch Virol. 1999;144(12):2312–2330.View this article via: PubMed Google Scholar
- Dejnirattisai W, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 2015;16(2):170–177.
- Wines BD, et al. The IgG Fc contains distinct Fc receptor (FcR) binding sites: the leukocyte receptors Fc gamma RI and Fc gamma RIIa bind to a region in the Fc distinct from that recognized by neonatal FcR and protein A. J Immunol. 2000;164(10):5313–5318.
- Zeisel MB, Baumert TF. Production of infectious hepatitis C virus in tissue culture: a breakthrough for basic and applied research. J Hepatol. 2006;44(2):436–439.
- Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796.
-
Version history
- Version 1 (May 19, 2022): In-Press Preview
- Version 2 (June 22, 2022): Electronic publication










