Research ArticleOncologyStem cells Free access | 10.1172/jci.insight.98096
Temporal DNA-PK activation drives genomic instability and therapy resistance in glioma stem cells
Yanling Wang,1 Haineng Xu,1 Tianrun Liu,1 Menggui Huang,1 Param-Puneet Butter,1 Chunsheng Li,2 Lin Zhang,2 Gary D. Kao,1 Yanqing Gong,3 Amit Maity,1 Constantinos Koumenis,1 and Yi Fan1,4
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Wang, Y. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Xu, H. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Liu, T. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Huang, M. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Butter, P. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Li, C. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Zhang, L. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Kao, G. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Gong, Y. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Maity, A. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Koumenis, C. in: PubMed | Google Scholar
1Department of Radiation Oncology,
2Department of Obstetrics & Gynecology,
3Division of Human Genetics and Translational Medicine, Department of Medicine, and
4Department of Neurosurgery, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
Find articles by Fan, Y. in: PubMed | Google Scholar
Published February 8, 2018 - More info
JCI Insight. 2018;3(3):e98096. https://doi.org/10.1172/jci.insight.98096.
Copyright © 2018, American Society for Clinical Investigation
Received: October 16, 2017; Accepted: December 28, 2017
-
Abstract
Cancer stem cells (CSCs) — known to be resistant to genotoxic radiation and chemotherapy — are fundamental to therapy failure and cancer relapse. Here, we reveal that glioma CSCs are hypersensitive to radiation, but a temporal DNA repair mechanism converts the intrinsic sensitivity to genomic instability and treatment resistance. Transcriptome analysis identifies DNA-dependent protein kinase (DNA-PK) as a predominant DNA repair enzyme in CSCs. Notably, DNA-PK activity is suppressed after irradiation when ROS induce the dissociation of DNA-PKcs with Ku70/80, resulting in delayed DNA repair and radiosensitivity; subsequently, after ROS clearance, the accumulated DNA damage and robust activation of DNA-PK induce genomic instability, facilitated by Rad50-mediated cell-cycle arrest, leading to enhanced malignancy, CSC overgrowth, and radioresistance. Finally, we show a requisite in vivo role for DNA-PK in CSC-mediated radioresistance and glioma progression. These findings identify a time-sensitive mechanism controlling CSC resistance to DNA-damaging treatments and suggest DNA-PK/Rad50 as promising targets for CSC eradication.
-
Introduction
Cancer stem cells (CSCs), also known as tumor-initiating cells or tumor-propagating cells, are highly tumorigenic and are capable of repopulating a tumor mass; importantly, CSCs are refractory to conventional cytotoxic radiation and chemotherapy (1–3). Accumulating evidence indicates that conventional treatments induce the enrichment of a CSC-like cell population within tumors, which leads to treatment resistance and, eventually, cancer relapse and metastasis. Therefore, new therapies that could effectively eradicate CSCs are urgently needed.
Glioblastoma multiforme (GBM), the grade IV glioma, is among the most lethal of human malignancies, with a current median survival of approximately 14 months (4). Most GBM tumors are highly resistant to treatment, and cancer recurrence is essentially universal, despite aggressive standard-of-care treatments, including surgical resection, radiation, and chemotherapy. Recent studies have identified a prominent CSC population, which is pluripotent and has the ability to repopulate tumors in brain tumors, including GBM (5–8). Glioma CSCs are resistant to radiotherapy and chemotherapy in vivo (9, 10), underscoring their role in therapy failure and cancer recurrence. Although strong clinical evidence supports an enrichment of CSC-like cells after treatment (11), heterogeneous responses for intrinsic radioresistance have been observed in patient-derived glioma CSCs (9, 12), implicating the existence of a complex regulatory mechanism.
The regulatory mechanism that controls DNA repair and therapy resistance in CSCs remains largely unknown. Checkpoint kinase 1/2–mediated (Chk1/2-mediated) high intrinsic DNA repair activity may induce radioresistance in glioma CSCs (9). Interestingly, a previously published study shows that skin bulge stem cells exhibit high DNA-dependent protein kinase (DNA-PK) activity relative to matched non–stem cells (13). More recent work suggests that DNA-PK is critical for treatment resistance in glioma CSCs (14, 15). However, how DNA-PK regulates DNA repair and radioresistance in CSCs is mainly unclear. Herein, we report a temporally regulated, DNA-PK–mediated mechanism that controls DNA repair in CSCs, results in treatment resistance in CSCs, and may serve as a promising therapeutic target for eliminating CSCs in cancer therapy.
-
Results
Time-dependent radioresistance is associated with delayed H2AX phosphorylation and DNA repair after radiation in CSCs. We investigated the responses of patient GBM tumor-derived CSCs to radiation and compared them to those of matched non–cancer stem cells (non-CSCs). CSCs and non-CSCs were cultured in same serum-free stem cell medium for analyses. Unexpectedly, during the first 1 week after irradiation, cell proliferation analyses showed that multiple lines of CSCs were consistently hypersensitive to radiation when compared with non-CSCs (differentiated, induced by serum) and glioma cell lines. Remarkably, despite being hypersensitive to radiation initially, the CSCs eventually regrew in the later phase, while the numbers of viable control cells were continuously reduced (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98096DS1), suggesting that a temporal mechanism may contribute to CSC radioresistance. These findings suggest that a time-sensitive mechanism may contribute to CSC radioresistance.
 Figure 1
Figure 1Time-dependent radioresistance is associated with increased DNA damage and delayed γ-H2AX and DNA repair after irradiation in glioma CSCs. (A) Human patient-derived IN528 CSCs and the matched non–cancer stem cells (non-CSCs) were cultured in serum-free stem cell medium and either irradiated by 5-Gy x-ray or were not irradiated. The number of viable cells was determined using cell viability assays. The data are presented as a percentage based on the number of viable nonirradiated cells of the same type (mean ± SEM, n = 3). (B) IN528 CSCs and matched non-CSCs were irradiated with 5-Gy x-ray. Cells were harvested at different times after irradiation. DNA damage was assessed using the single-cell gel electrophoresis comet assay with a neutral condition. Representative images and quantification of the percentage of DNA in the comet tails (mean ± SD). Scale bar: 50 μm. (C) IN528 CSCs and matched non-CSCs and glioma U87 cells were irradiated with 5-Gy x-ray. Cell lysates were immunoblotted with anti–P-H2AX-Ser139 (γ-H2AX) and anti-GAPDH antibodies. Representative blots. γ-H2AX band intensity was quantified and expressed as a percentage of its expression level before radiation. (D) CD133+ CSCs and CD133– non-CSCs were isolated from IN528 GBM cells and irradiated with 5-Gy x-ray. Cell lysates were immunoblotted with anti–P-H2AX-Ser139 (γ-H2AX) and anti-GAPDH antibodies. (E) IN528 CSCs and matched non-CSCs were irradiated with 5-Gy x-ray. Nuclear extracts were incubated with linearized DNA in NHEJ reaction buffer, followed by electrophoresis and gel imaging. The reaction mixtures were immunoblotted using an anti-PCNA antibody. Band intensity of multimers was analyzed.
In addition to the initial hypersensitivity to radiation and subsequent radioresistance in CSCs, DNA comet assays revealed a distinct pattern of DNA damage in CSCs following irradiation. Specifically, in contrast to the peak in DNA damage at 4 hours after irradiation in non-CSCs (similar dynamics have been extensively documented in a variety of somatic cell types), a continuously increasing DNA damage during 48 hours after radiation (Figure 1B), followed by a gradual decrease to the basal level in next 2–3 days (data not shown), was observed in CSCs. Moreover, irradiated CSCs showed a substantial delay in H2AX phosphorylation at Ser139 (γ-H2AX), a hallmark feature of DNA damage and repair (Figure 1C). CSCs, non-CSCs, and U87 glioma cells or primary GBM cells (data not shown) had different patterns of γ-H2AX that peaked at 72, 12, and 0.5 hours after irradiation, respectively. Similar results were observed in differentiated or undifferentiated multiple CSC lines derived from different patients (Supplemental Figure 2) and in CD133+ and CD133– cells in same patient (Figure 1D). Considering the major role of nonhomologous end joining (NHEJ) over that of homologous recombination for repairing double-stranded DNA breaks in multicellular eukaryotes, such as humans and mice (16), we analyzed NHEJ activity in irradiated CSCs. Consistent with the observation in γ-H2AX dynamics, DNA NHEJ analysis revealed robust, delayed DNA repair at 48 hours after irradiation in CSCs (Figure 1E). Together, these findings suggest that CSCs develop elongated DNA damage and delayed DNA repair after radiation, which may induce radiosensitivity during the initial phase.
DNA-PK is selectively required for H2AX phosphorylation after irradiation and DNA repair in CSCs. To explore potential molecular mechanisms underlying the temporal process of DNA damage and repair in CSCs, we utilized deep RNA sequencing (RNA-seq) to analyze the expression of DNA damage and repair-associated genes at the transcriptome level. Our gene set analysis of CSCs and matched non-CSCs revealed a stemness-associated upward trend of global gene expression, particularly in NHEJ, mismatch excision repair, and homologous recombination pathways (Figure 2A and Supplemental Figure 3). Consistent with the stronger NHEJ activities in CSCs before and after irradiation (Figure 1E), transcription of DNA-PK catalytic subunit (DNA-PKcs, PRKDC), a key enzyme that regulates NHEJ, was increased by 80% in CSCs (Figure 2A). Moreover, DNA-PKcs phosphorylation at Ser2056, a measure of its activation, was delayed after irradiation in CSCs and peaked at 48 hours (Figure 2B), which is consistent with the delayed DNA repair kinetics (Figure 1C). In contrast, the phosphorylation of ATM and ATR, two other key DNA repair enzymes, occurred rapidly after radiation (Figure 2B). These findings provide evidence that DNA-PKcs may play a selective role in regulating DNA repair in CSCs. Consistently, pharmacological inhibition of DNA-PK selectively and almost completely abolished radiation-induced γ-H2AX in CSCs but not in non-CSCs (Figure 2C). Likewise, knockdown of DNA-PKcs, but not ATM or ATR, remarkably abrogated radiation-induced γ-H2AX and DNA repair during the later phase (Figure 2, D and E) and induced more DNA damage (Figure 2F).
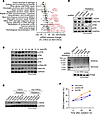 Figure 2
Figure 2DNA-PK is selectively required for after radiation γ-H2AX and DNA repair in CSCs. (A) RNA was isolated from IN528, T3961, and T4121 CSCs and from their corresponding matched non-CSCs and subjected to deep RNA-sequencing analysis. Approximately 200 genes encoding all known human proteins associated with DNA damage and repair were classified. Generated gene sets were ranked based on their mean percentage expression changes in CSCs versus non-CSCs and plotted as mean ± SEM. Genes with expression changes of more than 50% are annotated. (B) IN528 CSCs were irradiated with 5-Gy x-ray. Cell lysates were harvested at different time points after irradiation and subjected to immunoblot analysis. (C) IN528 CSCs and matched non-CSC cells were pretreated with 1 μM NU7441 (DNA-PK inhibitor), KU60019 (ATM inhibitor), or VE821 (ATR inhibitor) and irradiated by 5-Gy x-ray. At 48 hours after irradiation, cell lysates were collected and immunoblotted with anti–P-H2AX-Ser139 and anti-GADPH antibodies. (D–F) IN528 CSCs were transduced with lentivirus to express shRNAs targeting control GFP, DNA-PK, ATM, or ATR, followed by puromycin selection for stable expression cell lines. Cells were irradiated with 5-Gy x-ray. (D) Cells were harvested at 48 hours after irradiation. Cell lysates were subjected to immunoblot analysis. (E) Cells were harvested 48 hours after irradiation. Nuclear extracts were incubated with linearized DNA in NHEJ reaction buffer, followed by electrophoresis and gel imaging. The reaction mixtures were immunoblotted with anti-PCNA antibody. (F) Cells were harvested at different time points after irradiation. DNA damage was assessed using a single-cell gel electrophoresis comet assay. Quantification of the percentage of DNA in the comet tails is shown (mean ± SD).
DNA-PKcs is preferentially expressed in CSCs and is required for CSC survival after radiation. To understand the selective role of DNA-PKcs in DNA repair in CSCs, we examined the expression levels of key DNA repair enzymes, including DNA-PK, ATM, and ATR. RNA-seq revealed robust increases in DNA-PKcs in CSCs (Figure 3A). Immunoblot analysis verified stemness-associated DNA-PKcs expression in CSCs (Figure 3B). Importantly, CSCs showed robust, intrinsic DNA-PKcs expression, as indicated by 8-fold higher fragments per kilobase of transcript per million mapped reads (FPKM) values than ATM (Supplemental Figure 4A). Analysis of The Cancer Genome Atlas (TCGA) database showed a 3-fold increase in DNA-PK expression relative to ATM in GBM tumor cells (Supplemental Figure 4B), suggesting that DNA-PKcs may serve as a key regulatory enzyme for DNA repair in CSCs. Consistent with its preferential expression in CSCs, knockdown of DNA-PKcs, or, to a lesser extent, ATM, markedly sensitized radiation-induced cytotoxicity in multiple lines of CSCs (Figure 3C and Supplemental Figure 5) but not in non-CSCs (Figure 3D).
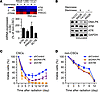 Figure 3
Figure 3DNA-PK is preferentially expressed in CSCs and is critical for after radiation survival. (A) RNA was isolated from IN528, T3961, and T4121 CSCs and their matched non-CSCs and subjected to deep RNA-sequencing analysis. Heat map of the expression values for different genes and quantified percentages of each expressed gene (CSCs versus non-CSCs, mean ± SEM, n = 3). (B) IN528 and T4121 CSCs and their matched non-CSCs were subjected to immunoblot analysis. (C) IN528 CSCs and (D) matched non-CSCs were transduced with lentivirus that express shRNAs targeting GFP, DNA-PK, or ATM, followed by puromycin selection for stable expression cells. Cell were either irradiated 5-Gy x-ray or were not irradiated. The number of viable cells was determined using cell viability assays. The data are presented as a percentage based on the number of viable nonirradiated cells of the same type (mean ± SEM, n = 3).
ROS induces temporal DNA-PKcs inhibition by disrupting Ku70/80 binding to DNA-PKcs, leading to delayed H2AX phosphorylation and DNA repair. We further explored the regulatory mechanisms for DNA-PKcs–mediated DNA repair in CSCs. Consistent with previous reports showing the robust ability of CSCs to scavenge ROS (17), our data showed more rapid clearance of peroxide hydrogen (H2O2) and total ROS in CSCs than in non-CSCs (Figure 4A and Supplemental Figure 6). Interestingly, ROS concentration reached the basal level 24 hours after irradiation, which coincided with DNA-PK activation. To test whether ROS regulates DNA-PKcs activity, CSCs were pretreated with the ROS scavenger TEMPOL, followed by x-ray irradiation. Our data showed that scavenging ROS with TEMPOL remarkably altered DNA-PKcs phosphorylation, shifting the peak phosphorylation from 48 hours to 6 hours and diminishing DNA-PKcs activation after 12 hours following irradiation, although it did not affect ATM phosphorylation (Figure 4B). Interestingly, TEMPOL pretreatment substantially restored the interaction between DNA-PKcs and Ku70/80, which is known to induce DNA-PK autophosphorylation and activation, before and, to a greater extent, after radiation (Figure 4C), suggesting that ROS regulates DNA-PKcs binding to Ku70/80, leading to the inhibition of its activity. Furthermore, ROS scavenging remarkably accelerated the appearance of γ-H2AX (Figure 4D) and abrogated the DNA repair at 48 hours after irradiation (Figure 4E), most likely due to suppressed DNA-PK activity at that time (Figure 4B).
 Figure 4
Figure 4ROS induces time-dependent DNA-PK inhibition by disrupting Ku70/80 binding to DNA-PKcs, leading to delayed H2AX phosphorylation and DNA repair. (A) IN528 CSCs and matched non-CSCs were irradiated with 5-Gy x-ray. Intracellular ROS levels (H2O2) were measured by luminol-based luminescence analysis at different time points after irradiation (mean ± SEM, n = 3). (B–E) IN528 CSCs were pretreated with TEMPOL (10 mM) and irradiated with 5-Gy x-ray. (B) Cells were harvested at different time after irradiation and were subjected to immunoblot analysis. (C) At 4 hours after irradiation, cell lysates were immunoprecipitated with anti–DNA-PKcs or anti-Ku80 antibodies, followed by immunoblot analysis with anti-Ku80 or anti–DNA-PKcs antibody. (D) Cells were harvested at different times after irradiation and analyzed by immunoblot with anti–P-H2AX-Ser139 (γ-H2AX) and anti-GAPDH antibodies. (E) Cells were harvested different time after radiation. Nucleic extracts were incubated with linearized DNA in NHEJ reaction buffer, followed by electrophoresis and gel imaging. The reaction mixtures were immunoblotted with anti-PCNA antibody.
Delayed DNA-PK activation induces genomic instability and enhances cell malignancy in CSCs. Accumulation of DNA damage is known to induce genomic instability, leading to cell transformation and tumorigenesis. Therefore, we hypothesized that delayed DNA-PK activation and DNA repair may induce genomic instability in CSCs, driving cell regrowth and radioresistance. To test our hypothesis, we determined the effects of radiation on genomic stability in CSCs and non-CSCs. Our data showed that radiation induced the formation of a micronucleus, a marker of genomic instability, in CSCs, whereas an approximately 40% decrease in micronucleus formation was observed in irradiated non-CSC cells (Figure 5A). Furthermore, flow cytometric analysis showed that radiation induced a gradual increase in chromosomal aneuploidy in CSCs; in contrast, radiation induced half as much aneuploidy in non-CSC cells (Figure 5, B and C), suggesting a critical role for stemness in radiation-induced genomic instability.
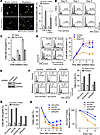 Figure 5
Figure 5Delayed DNA-PK activation induces genomic instability and enhances cell malignancy in CSCs. (A–C) IN528 CSCs and matched non-CSCs were either irradiated with 5-Gy x-ray or were not irradiated (sham control). (A) At 2 days after irradiation, the cells were subjected to micronucleus analyses. Representative images and quantification of data (mean ± SD, n = around 1,000 cells). Scale bar: 50 μm. (B and C) Cells were analyzed by flow cytometry for aneuploidy. (B) Representative sorting. (C) Quantified data (mean ± SEM, n = 3). (D) IN528 CSCs were pretreated with 1 μM KU60019 (inhibitor of ATM [iATM]), NU7441 (inhibitor of DNA-PK [iDNA-PK]), or control 0.1% DMSO, and irradiated with 5-Gy x-ray. At different times after irradiation, cells were subjected to aneuploidy analysis by flow cytometry (mean ± SEM, n = 3). (E–I) IN528 CSCs were transduced with lentivirus that expresses shRNAs targeting control GFP, DNA-PK, Rad50, or ATM, followed by puromycin selection for stable expression cell lines. Cells were irradiated by 5-Gy x-ray. (E) Cells were analyzed by immunoblot. (F) 24 hours after irradiation, cells were stained with propidium iodide and analyzed by flow cytometry for cell cycle. Representative sorting and quantitative results (mean ± SEM, n = 3). (G) 72 hours after irradiation, cells were stained with propidium iodide and analyzed by flow cytometry for cell aneuploidy (mean ± SEM, n = 3). (H) Cells were subjected to viability analysis at different time points after radiation (mean ± SEM, n = 3-6). (I) At 2 weeks after irradiation, cells were subjected to neurosphere formation/survival fraction analysis (mean ± SEM, n = 3–5).
To test the role of DNA-PK in radiation-induced chromosomal instability, CSCs were pretreated with specific pharmacological inhibitors of DNA-PK or ATM, followed by radiation. Our data showed that inhibition of DNA-PK, but not ATM, markedly inhibited radiation-induced aneuploidy in CSCs (Figure 5D) and neither inhibition substantially affected radiation-induced aneuploidy in non-CSCs (Supplemental Figure 7). In addition, we also tested the role of Rad50 in cell cycle and aneuploidy regulation, which was identified as an overexpressed DNA repair-associated gene by our RNA-seq analysis (Figure 2A). Our data showed that shRNA-mediated knockdown of Rad50, but not DNA-PK, inhibited radiation-induced G2/M arrest of cell cycle in CSCs (Figure 5, E and F). Moreover, knockdown of DNA-PK or Rad50, but not ATM, inhibited radiation-induced aneuploidy in CSCs (Figure 5G), implicating that Rad50 may facilitate the delayed DNA-PK activation-mediated genomic instability by regulating G2 cell cycle arrest. Furthermore, DNA-PKcs knockdown, and, to a lesser extent, Rad50 knockdown, abolished the CSC regrowth 7 days after irradiation (Figure 5H) and abrogated cell survival after irradiation (Figure 5I). Together, these findings suggest that DNA-PK–mediated temporal regulation of DNA repair is critical for radiation-induced genomic instability in CSCs.
DNA-PK is critical for genomic instability and radioresistance in CSCs in vivo. To rigorously determine the role of DNA-PK in maintaining genomic instability and CSC radioresistance in vivo, we induced orthotopic gliomas in a xenograft nude mouse model by using human CSCs, followed by CT-guided stereotactic radiotherapy (Figure 6A). Our data showed that low-dose (3-Gy) radiation did not significantly improve animal survival in control mice (Figure 6B). However, either DNA-PKcs or ATM knockdown sensitized CSC-derived gliomas to radiation, leading to increased mouse survival. Notably, around 30% of the irradiated mice with DNA-PKcs–knockdown tumors survived after all of the other mouse groups had died (at day 70), and 10% of these mice survived for at least 90 days, when the experiment was terminated (Figure 6B). Consistent with the critical role of DNA-PKcs in mouse survival, knockdown of DNA-PKcs, but not ATM, plus radiation significantly reduced tumor growth, as indicated by a 65% decrease in average tumor volume (at day 41 after tumor implantation) (Figure 6C). Furthermore, DNA-PK knockdown robustly enhanced radiation-induced tumor necrosis and increased cell apoptosis, as shown by TUNEL staining (Figure 6, D and E). Importantly, knockdown of DNA-PKcs, but not ATM, efficiently abrogated radiation-induced genomic instability, as indicated by a marked reduction in aneuploid cell populations in the tumors (Figure 6F). Together, these data indicate that DNA-PK plays a critical role in radioresistance and radiation-induced genomic instability in CSCs in vivo.
 Figure 6
Figure 6DNA-PK is critical for genomic instability and radioresistance in CSCs in vivo. Human IN528 CSCs that stably express luciferase and shRNAs targeting control GFP, ATM, or DNA-PK were injected into the brains of nude mice. Using guided micro-CT, mice were locally irradiated with stereotactic x-ray radiation (3 Gy, single dose). (A) Experimental procedure. (B) Animal survival was monitored for 90 days after injection (n = 7–9 mice). P values were determined by log-rank (Mantel-Cox) tests. MS, median survival. (C) Tumor growth was analyzed by bioluminescence. Representative images and quantitative analysis of integrated luminescence in tumors at day 41 (mean ± SEM). P values were determined by t tests. (D) Tumor sections were stained with H&E. Representative images are shown (n = 4–5 mice). Scale bar: 100 μm. (E) Tumor sections were stained by TUNEL. Representative images are shown, and TUNEL+ cells were counted (mean ± SEM, n = 3 mice). Scale bar: 100 μm. (F) Single-cell suspensions derived from tumors were stained with propidium iodide and subjected to aneuploid analysis by flow cytometry. Representative sorting and quantified data (mean ± SEM, n = 4–9 mice) are shown. P values were determined by t tests.
Pharmacological inhibition of DNA-PK in GBM-bearing mice overcomes CSC-mediated radioresistance and leads to enhanced survival. Finally, we tested an experimental therapy for CSC eradication by utilizing a genetic GBM mouse model that recapitulates major pathological features of human GBM (Figure 7A). Fractioned high-dose (10-Gy) radiation significantly improved mouse survival by 9 days, whereas pharmacological inhibition of ATM or DNA-PK alone did not increase survival (Figure 7B). Importantly, DNA-PK inhibition induced significant sensitization to radiotherapy, resulting in a nearly doubled overall animal survival (from 22 days to 41.5 days). In contrast, ATM inhibition moderately improved the radiotherapy (from 22 days to 34 days). Moreover, radiotherapy plus inhibition of DNA-PK, but not inhibition of ATM, significantly reduced tumor growth, as indicated by a 70% decrease in average tumor volume (at day 24 after tumor implantation) (Figure 7C). Furthermore, radiotherapy plus DNA-PK inhibition, but not ATM inhibition, efficiently eradiated the CD133+ CSC population (Figure 7, D and E). In addition, radiotherapy plus DNA-PK inhibition suppressed tumor progression and abrogated hemorrhagic necrosis, a defining pathological feature of GBM (Figure 7F), likely due to the eradication of CSCs. Together, these results provide evidence that radiotherapy plus DNA-PK inhibition may be an efficient approach for eradicating CSCs in gliomas.
 Figure 7
Figure 7Combined radiotherapy with DNA-PK inhibition eradicates CSCs and improves mouse survival. Primary GBM in Ntv-a;Ink4a–/–;Ptenfl/fl;LSL-Luc donor mice is induced by RCAS-mediated somatic gene transfer. Recipient mice were wild-type B6 mice. Mice were treated with 10 mg/kg NU7441 (iDNA-PK) or KU60019 (iATM) for 8 days and simultaneously irradiated with x-ray (2.5-Gy doses, 4 times). (A) Experimental procedure. (B) Animal survival was monitored for 65 days after injection (n = 10–11 mice). P values were determined by log-rank (Mantel-Cox) tests. MS, median survival. (C) Tumor growth was analyzed by bioluminescence. Representative images and quantitative analysis of integrated luminescence in tumors at day 24 (mean ± SEM). P values were determined by t tests. (D and E) Tumor sections were probed with anti-CD133 antibody and counterstained with hematoxylin. (D) Representative images. Scale bar: 100 mm. (E) Quantitative results (mean ± SD, n = 5 mice). (F) Tumor sections were stained with H&E. Representative images are shown (n = 4–5 mice). Scale bar: 100 μm.
Collectively, these findings reveal a time-dependent DNA repair mechanism that drives treatment resistance in CSCs (Figure 8). Specifically, DNA-PKcs activity is suppressed after irradiation by ROS-induced dissociation of Ku70/80, leading to delayed DNA repair and radiosensitivity. Subsequently, following ROS clearance, the accumulated DNA damage accumulation and the robust reactivation of DNA-PK induce genomic instability, resulting in cell malignancy and eventually leading to CSC overgrowth and radioresistance. In addition, Rad50 promotes cell cycle G2 arrest, facilitating the DNA-PK–mediated processes.
 Figure 8
Figure 8Schematic model. A schematic model for DNA-PK–mediated temporal control of radioresistance in CSCs.
-
Discussion
It is increasingly recognized that CSCs are a major driver of treatment resistance and cancer recurrence and metastasis. Multiple mechanisms contribute to the resistance of CSCs to treatment, including cell dormancy, increased drug efflux and detoxification, activation of antiapoptotic signal pathways, robust abilities to scavenge ROS, and enhanced activities for DNA repair (17, 18). Notably, previous studies suggest that high intrinsic DNA repair activity — mediated by Chk1/2 — drives radioresistance in glioma CSCs (9). Our RNA-seq–based findings, which show that CSCs exhibit a global increase in the expression of most DNA repair-associated genes, seem to support this prevailing theory. However, we also discovered that glioma CSCs are hypersensitive to radiotherapy during the initial phase (as evidenced by more DNA damage and less cell viability after irradiation), which is consistent with more recent studies showing radiosensitivity in multiple lines of patient-derived glioma CSCs (12), suggesting that treatment resistance may be time dependent in different CSCs. More importantly, our study reveals a DNA-PK–mediated mechanism that temporally regulates DNA repair and leads to treatment resistance by inducing genomic instability in CSCs (Figure 8), suggesting that radioresistance in CSCs is not simply driven by intrinsically enhanced DNA repair.
H2AX phosphorylation at Ser139 (γ-H2AX) is a hallmark feature of DNA damage and repair (19, 20). Rapid induction of γ-H2AX, which peaks at 0.5–2.0 hours after irradiation, has been extensively documented in a variety of somatic cell types. Unexpectedly, our data revealed that γ-H2AX induction peaks at 24–48 hours after irradiation in CSCs, providing evidence for delayed DNA repair in CSCs. This delay in DNA repair, combined with an increased period of DNA damage, may provide a window of opportunity for CSCs to accumulate DNA damage, which eventually results in genomic instability. NHEJ occurs predominantly in G1 and G2 during the cell cycle (21). Likewise, the robust activation of DNA-PKcs during the later phase, likely facilitated by Rad50-dependent G2 arrest in cell cycle, induces NHEJ DNA repair and allows cells to reenter the cell cycle and to proliferate. Subsequently, their genomic instability induces oncogenic transformation and promotes cell division in viable CSCs. In addition, DNA-PK–independent modes of NHEJ may exist at the early phase, considering that cancer cells also repair their damaged DNA via alternative NHEJ. These findings identify a time-dependent mechanism that controls radioresistance in CSCs.
Our findings suggest that CSCs selectively utilize DNA-PK to repair their damaged DNA, likely due to the constitutively high expression of DNA-PKcs and relatively low expression of other DNA repair enzymes, including ATM and ATR, as revealed by our comparative RNA-seq analysis. Notably, a recent report shows that skin bulge stem cells exhibit high DNA-PK activity relative to matched non–stem cells (13). This finding is consistent with our results showing stemness-associated DNA-PK expression. Specifically, DNA-PKcs is preferentially expressed in CSCs relative to non-CSCs, suggesting that CSCs may utilize a different DNA repair mechanism in response to therapy-induced DNA damage.
Furthermore, our study reveals a previously unidentified regulatory system capable of controlling DNA-PK activity, in which ROS regulates DNA-PK activity by modulating DNA-PKcs’ interaction with Ku70/80, a key process that induces DNA-PKcs autophosphorylation and activation (22–24). Radiation induces an initial increase in ROS generation, likely due to early direct radiation effects (water radiolysis and OH production) and later indirect mitochondrial dysfunction, followed by a gradual decrease in ROS generation that is likely induced by highly expressed free radical scavenging systems, e.g., glutathione (GSH) biosynthesis, in CSCs (17). These ROS dynamics temporally regulate DNA-PKcs activity, along with the preferential expression of DNA-PKcs and the selective requirement of DNA-PKcs for DNA repair, which serve as dual mechanisms that lead to the delayed, robust activation of DNA-PKcs and NHEJ in CSCs during the later time phase after irradiation.
We report a unique mechanism by which CSCs develop increased malignancy and treatment resistance achieved through enhanced genomic instability. Specifically, the accumulation of DNA damage combined with temporally regulated NHEJ repair could result in a high number of genetic mutations in CSCs, which leads to genomic instability and fuels oncogenic transformation. These findings provide insights into the clinical observation of enhanced malignancy in relapsed tumors that likely are derived from treatment-transformed CSCs. Likewise, genomic instability is a common feature observed during the later stages of primary tumor development or in metastasized tumors at distant sites (25, 26), both of which are highly associated with CSCs (27, 28). In addition, we also expect that genotoxic chemotherapy, e.g., with temozolomide and doxorubicin that damage DNA, may have similar effects on genomic instability in CSCs as radiotherapy. Our data established requisite in vitro and in vivo roles for DNA-PK in maintaining genomic instability in CSCs. In contrast, previous studies show that DNA-PKcs–knockout mice exhibit increased spontaneous and radiation-induced genomic instability in fibroblasts (29), suggesting that this stemness-associated mechanism may strictly depend on the preferential expression of DNA-PKcs over ATM and ATR, the temporally regulated activity of DNA-PKcs, and/or the altered global expression profile of DNA repair-associated genes in CSCs.
Pharmacological inhibition of DNA damage response enzymes, including DNA-PK, ATM, and ATR, has emerged as a crucial strategy for sensitizing tumor cells to radiotherapy and chemotherapy over last two decades (30–32). Recent preclinical studies using these inhibitors show promising results; however, their effects on the sensitization appear marginal to moderate (33–36), possibly due to redundant functions among these kinases and other compensatory mechanisms (30, 31, 37). Previous work also suggests a role of DNA-PK for therapeutic resistance in cancer stem cells by regulating autophagy, expression of multidrug resistance genes, and DNA repair (14, 15, 38). Our results reveal a spatial regulation mechanism and provide evidence that DNA-PK is selectively required for the DNA damage response in glioma CSCs. In this study, we show that DNA-PK inhibition by NU7441, a potent and selective DNA-PK inhibitor (39), eliminates the CSC population and improves the survival of mice bearing gliomas. This result could be subjected to further improvement, considering the limited aqueous solubility and in vivo rapid clearance of NU7441 (31). Interestingly, mice lacking DNA-PKcs develop normally, but humans with null mutations are likely not viable (40). The unique requirement for DNA-PKcs in human cells may be a clinical barrier to developing DNA-PK as a therapeutic target. Thus, development of new and improved pharmacological or genetic approaches that efficiently and sustainably inhibit DNA-PK activity will contribute to CSC-targeted anticancer treatments, which, when combined with additional cytotoxic and targeted therapies, may achieve maximal therapeutic efficacy in cancer intervention.
In summary, our study reveals a time-dependent, DNA-PK–mediated mechanism controlling treatment resistance in glioma CSCs mediated through genomic instability. Furthermore, genetic ablation and pharmacological inhibition of DNA-PK abrogate CSC growth, inhibit glioma progression, and sensitize tumors to radiotherapy in vivo. Thus, targeting DNA-PK may provide an unprecedented opportunity to eliminate CSCs, overcome treatment resistance, and block cancer relapse.
-
Methods
Cell culture. Human patient-derived IN528, T3565, T3961, and T4121 glioma CSCs were harvested, characterized, and provided by Jeremy Rich (Cleveland Clinic, Cleveland, Ohio, USA) (9, 41, 42). The stemness of these cells was previously validated by stem cell marker expression (CD133, OLIG2, and SOX2), functional assays of self-renewal (serial tumorsphere passage), and in vivo tumor formation assays (41, 43). The matched non-CSCs were generated by brief treatment with medium containing 10% FBS for 24 hours and then were put back into stem cell medium as described previously (9, 44, 45). As an additional control, human GBM patient-derived IN528 cells were incubated with biotinylated anti-CD133 antibody (Miltenyi Biotec, 130090664), and CD133+ and CD133– cells were harvested by magnetic-activated cell sorting with streptavidin-conjugated microbeads (Miltenyi Biotec, 130048101). CSCs were cultured in serum-free Neurobasal-A medium (Gibco), supplemented with B-27 Supplement Minus Vitamin A (Gibco), GlutaMax (Gibco), sodium pyruvate (Gibco), fibroblastic growth factor (5 ng/ml, R&D Systems), and epidermal growth factor (20 ng/ml, R&D Systems). Culture medium was changed every 2 days. Cells were maintained and limited to a low number of passages (less than 5).
Cell irradiation. Cells were treated by irradiation by x-ray with a dose rate of 2.8 Gy/min at room temperature using a X-Rad 320ix cabinet system (Precision X-Ray). Cells were pretreated with 10 mM ROS scavenger 4-Hydroxy-TEMPO (TEMPOL, MilliporeSigma) and irradiated with single-dose (5-Gy) irradiation.
Cell viability assay. Cells were trypsinized, and the single-cell suspensions were loaded into white 96-well plates, followed by overnight culture and different treatments. Cell viability was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, G7571) according to the manufacturer’s instructions. Luminescence was detected using a Synergy H4 Hybrid luminescent plate reader (BioTek).
Clonogenic/sphere-forming assay. CSCs were trypsinized, and the single-cell suspensions were loaded into 96-well plates. Cells were cultured overnight and subjected to different treatments. After the cells were incubated at 37°C for 14 days, the number of neurospheres in CSCs was counted (46).
shRNA lentiviral transduction. The oligonucleotides (sense, 5′-GATCCCGGGCGC TAATCGTACTGAATTCAAGAGATTCAGTACGATTAGCGCCCTTTTTTGGAAA-3′, antisense, 3′-GGCCCGCGATTAGCATGACTTAAGTTCTCTAAGTCATGCTAATCGCGGGAAAAAACCTTTTCGA-5′) that encode small-hairpin RNAs (shRNAs) targeting DNA-PKcs (nucleotides 11637–11655) were synthesized and cloned into the pLV lentivirus expression vector by VectorBuilder (47). Lentiviral expression vectors that encode shRNAs targeting ATM or ATR were obtained from Addgene (14542 and 14543). The oligonucleotides (sense, 5′-TGGGATTCAATGTTCATTAATTCAAGAGATTAATGAACATTGAATCCCTTTTTTC-3′, antisense, 5′-TCGAGAAAAAAGGGATTCAATGTTCATTAATCTCTTGAATTAATGAACATTGAATCCCA-3′) that encode shRNAs targeting Rad50 were synthesized and cloned into the pSicoR lentivirus expression vector. To prepare the lentivirus, 293T cells were cotransfected with lentiviral expression vectors and packaging vectors (System Biosciences) for 8 hours. The medium was replaced with fresh medium, the cultures were incubated for 48 hours, and the medium supernatants containing lentivirus were collected. CSCs were infected with lentivirus in the presence of 8 mg/ml polybrene (MilliporeSigma). Stable shRNA-expressing cells were selected with 0.5 μg/ml puromycin and were maintained in the culture medium with 0.2 μg/ml puromycin.
Single-cell gel electrophoresis comet assay. Cells were subjected to neutral comet assays for detecting DNA double-strand breaks as described previously (48). In brief, after irradiation, cells were washed with PBS, mixed with low-melting agarose (1:10), and loaded into slides. The slides were then immersed in lysis solution (pH 8.0, 30 mM EDTA, 0.5% sodium dodecyl sulfate) overnight at 37°C. After lysis, slides were rinsed 3 times with 90 mM Tris buffer (pH 8.5) containing 90 mM boric acid and 2 mM EDTA and then subjected to horizontal electrophoresis at a rate of 0.6 V/cm for 25 minutes. DNA was stained with propidium iodide (BD Pharmingen) and imaged with an AxioImager A1 fluorescence microscope (Zeiss) equipped with an AxioCam 506 CCD camera (Zeiss). The cell number with DNA comet assay and the DNA percentage content in the comet tail region were measured using NIH ImageJ and OpenComet 1.3 software.
Deep RNA-seq analysis. IN528, T3961, and T4121 CSCs and matched non-CSCs were cultured and treated with CSC culture medium for 12 hours. Approximately 2 × 106 cells were lysed in 1 ml TRIzol (Thermo Fisher), followed by RNA extraction according to the manufacturer’s instructions. The isolated RNA was purified by using an RNeasy Plus Mini Kit (Qiagen). After a quality control step using RNA Nano assay chips with a 2100 bioanalyer (Agilent), the library was constructed by using a RiboZero protocol with TapeStation (Agilent) and subjected to deep sequencing (125 PE, about 40 megabyte reads for each sample, Illumina sequencer 2500) in the Next-Generation Sequencing Core at the University of Pennsylvania. Data analysis was performed in the core facility, and gene expression was expressed as FPKM values. About 200 genes encoding all known human proteins associated with DNA damage and repair were classified (http://sciencepark.mdanderson.org/labs/wood/dna_repair_genes.html) and matched with the FPKM expression profile (49–51). RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus under the accession GSE108322.
ROS assay. Cells were trypsinized, seeded into white 96-well plates, and cultured overnight, followed by x-ray irradiation. ROS (H2O2) levels were determined using the ROS-GLO H2O2 assay (Promega, G8820) according to the manufacturer’s instructions. Luminescence was measured using a Synergy H4 Hybrid luminescent plate reader (BioTek). Total ROS levels were measured using a Total Reactive Oxygen Species Assay Kit (eBioscience, 88593174). Cells were analyzed using an Accuri C6 flow cytometer (BD Biosciences) with FlowJo software.
Immunoblot analyses. Cells were lysed in cell lysis buffer (Cell Signaling, 9803) containing a complete protease inhibitor cocktail (1:1,000, Roche) and Halt phosphatase inhibitor cocktail (1:100, Thermo Fisher). 20 μg total protein was resolved by 4%–15% SDS-PAGE (Bio-Rad) and transferred to PVDF membranes. Membranes were blocked with 5% BSA and incubated with anti–phospho-H2AX-Ser139 (1:2,000, Cell Signaling, 9713), anti–phospho-DNA-PKcs-Ser2056 (1:1,000; Abcam, ab18192), anti–DNA-PKcs (1:1,000; Cell Signaling, 12311), anti–phospho-ATM-Ser1981 (1:1,000, Abcam, ab81292), anti-ATM (1:1,000; Cell Signaling, 2873), anti–phospho-ATR-Ser428 (1:1,000, Cell Signaling, 2853), anti-ATR (1:1,000, Cell Signaling, 13934), anti-PCNA (1:2,000, Cell Signaling, 2586), anti-Rad50 (1:1,000, Cell Signaling, 3427), and anti-GAPDH (1:5,000, Cell Signaling, 2118) antibodies, followed by incubation with a secondary antibody conjugated to horseradish peroxidase (1:2,000; Bio-Rad). Signals were visualized using ECL Prime Western Blotting Detection reagent (Amersham).
Coimmunoprecipitation analysis. IN528 CSCs were pretreated with 10 mM ROS scavenger 4-Hydroxy-TEMPO (TEMPOL, MilliporeSigma) and irradiated with 5-Gy x-ray. At 4 hours after irradiation, the cells were lysed in coimmunoprecipitation buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 μg/ml leupeptin). After centrifugation at 15,000 g for 15 minutes, 0.5 μg anti-Ku80 antibody (Thermo Fisher, MA5-12933), anti–DNA-PKcs antibody (Santa Cruz Biotechnology, sc9051), or control IgG (Santa Cruz Biotechnology, sc2027 and sc2025) was added to the supernatants (200 μg total protein), followed by an overnight incubation at 4°C. Protein A/G–conjugated agarose beads (Santa Cruz Biotechnology, sc2003) were added to each sample and incubated at 4°C for 2 hours, followed by centrifugation at 1,000 g for 10 minutes. The pellets were washed 4 times with PBS buffer and subjected to immunoblot analysis.
DNA aneuploidy and cell cycle assay. DNA aneuploidy was analyzed by flow cytometry as previously descried (52, 53). In brief, irradiated cells were washed with PBS and fixed with 70% ethanol. Cells were stained with propidium iodide (BD Pharmingen) in the presence of DNase-free RNase A (0.2 mg/ml, Thermo Fisher). Cells were analyzed using an Accuri C6 flow cytometer (BD Biosciences), and the data were analyzed using FlowJo software.
In vitro NHEJ assay. Plasmid-based assays for DNA end joining were performed as described previously (54). Briefly, nuclear proteins were harvested from irradiated IN528 cells by using a Nuclear Extraction Kit (Signosis). 10 μg extracted nuclear protein was incubated with 100 ng BamH1-linearized pEGFP-C1-FLAG plasmid (4,766 bp, Addgene, 46956) in 20 μl reaction buffer (20 mM HEPES, pH 7.5, 80 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT) for 1 hour at 25°C. The reactions were terminated by the addition of stop solution (1 μl of 10 mg/ml proteinase K, 2 μl of 0.5 M EDTA, pH 7.5, and 2 μl of 0.5% SDS) and incubated for 30 minutes at 37°C. The DNAs were resolved by agarose gel electrophoresis, followed by ethidium bromide staining. Band intensity of multimers was analyzed by using the NIH ImageJ program. In addition, the nuclear protein was immunoblotted with an anti-PCNA antibody (1:2,000, Cell Signaling, 2586).
Micronucleus analysis. The cytokinesis-block micronucleus cytome assay was performed as described previously (55). Briefly, irradiated cells were treated with 4 μg/ml cytochalasin B (MilliporeSigma). The cells were fixed with methanol/acetic acid (3:1, v/v) and stained with DAPI. Scoring of micronuclei was limited to binucleated cells with preserved cytoplasm. The results are expressed as the numbers of the cells with micronuclei per 1,000 binucleated cells.
Mice. Six- to eight-week-old, half male and half female, nude mice and wild-type black B6 mice were used (Jackson Laboratory).
Xenograft glioma model. IN528 CSCs were transduced with lentivirus to express firefly luciferase and an shRNA targeting DNA-PK or ATM, followed by puromycin selection. 2 × 104 cells in a total volume of 2 μl were orthotopically injected into nude mice by using a stereotaxic injection system (Stoelting). Mice were administrated with a focused radiation therapy with a single dose of 3-Gy x-ray, guided by micro-CT in a small animal radiation research platform. Tumor growth was monitored by whole-body bioluminescence using an IVIS 200 Spectrum Imaging System after retro-orbital injection of luciferin (150 mg/kg, GoldBio). Mice were euthanized when they exhibited severe GBM symptoms, including dome head, hemiparesis, or more than 20% of body weight loss. After irradiation, survival was monitored for 90 days.
Genetic glioma model. GBM was induced by RCAS-PDGF/Cre–mediated somatic gene transfer in Ntv-a;Ink4a-Arf−/−;Ptenfl/fl;LSL-luc mice as described previously (56–59). Tumors in the donor mice were excised and subjected to mechanical dissociation using a gentleMACS Dissociator (Miltenyi Biotec) and enzymatic digestion with collagenase II (Invitrogen) and dispase (Invitrogen) to obtain single-cell suspensions. Wild-type B6 mice were orthotopically, stereotactically injected with 2 × 105 GBM tumor cells. Tumor growth was monitored by whole-body bioluminescence using an IVIS 200 Spectrum Imaging System after retro-orbital injection of luciferin (150 mg/kg, GoldBio). At 11 days after injection, the mice were treated peritoneally with 10 mg/kg NU7441 (an inhibitor of DNA-PK, SelleckChem) or KU60019 (an inhibitor of ATM, SelleckChem) or control DMSO twice daily for 8 days. The mice were also administrated by focused radiotherapy with a single dose of 2.5 Gy every other day for 4 times using an X-Rad 320ix cabinet system (Precision X-Ray). Mice were euthanized when they exhibited severe GBM symptoms, including dome head, hemiparesis, or more than 20% of body weight loss. After treatment, survival was monitored for 65 days.
Histology and immunohistochemistry. Paraffin sections of mouse tumors were deparaffinized, rehydrated, and subjected to antigen retrieval in Target Retrieve Solution (Dako) at 95°C for 20 minutes. Tissue sections were stained with H&E. Tissues were imaged with an AxioLab microscope (Zeiss) equipped with an AxioCam HRC CCD camera (Zeiss). For immunohistochemistry, paraffin sections were deparaffinized and rehydrated, subjected to antigen retrieval in Target Retrieve Solution at 95°C for 20 minutes, and blocked with 5% horse serum for 1 hour at room temperature. Sections were incubated with anti-CD133 (1:100, Biolegend, 141202) antibody overnight at 4°C. Sections were incubated with biotinylated goat anti-rat IgG (1:200, Vector Laboratories, BA9401), followed by staining with a VECTASTAIN ABC-HRP Kit (Vector Laboratories, PK7200) according to the manufacturer’s instructions.
TUNEL staining. Paraffin sections of mouse tumors were deparaffinized and rehydrated and subjected to in situ apoptosis detection using the Click-iT Plus TUNEL Assay (Life Technology, C10618). Images were acquired with an AxioImager A1 fluorescence microscope (Zeiss) equipped with an AxioCam 506 CCD camera (Zeiss).
Statistics. Student’s t (unpaired 2-tailed analysis) and log-rank tests (Mantel-Cox analysis) were used for statistical analyses between groups using Prism software, and P values less than 0.05 were considered statistically significant.
Study approval. All experiments with mice were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania and with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). All animals were housed in the Association for the Assessment and Accreditation of Laboratory Animal Care–accredited animal facility of the University of Pennsylvania.
-
Author contributions
YW designed, performed, and analyzed experiments and produced figures. HX prepared RNA and performed the RNA-seq analysis. TL contributed to animal studies. MH and PPB contributed to immunofluorescence analyses. CL and LZ contributed to flow cytometry and RNA-seq analysis. GDK, YG, AM, and CK provided suggestions for experimental design. YF designed and supervised experiments and wrote the manuscript. All authors commented on the manuscript.
-
Acknowledgments
We are grateful to Jeremy Rich and Eric Holland for providing glioma CSCs and the RCAS-PDGF GBM model, respectively; to Shipa Rao for bioinformatical analyses of RNA-seq data; and to Eric Brown for helpful discussions. This work was supported in part by NIH grants R00HL103792 and R01NS094533 (to YF) and R01CA190415 (to LZ).
Address correspondence to: Yi Fan, Department of Radiation Oncology, University of Pennsylvania Perelman School of Medicine, 3400 Civic Center Boulevard, SCTR 8-132, Philadelphia, Pennsylvania 19104, USA. Phone: 215.898.9291; Email: fanyi@uphs.upenn.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(3):e98096. https://doi.org/10.1172/jci.insight.98096.
-
References
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111.View this article via: PubMed Google Scholar
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768.
- Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13(10):727–738.
- Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996.
- Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401.View this article via: PubMed Google Scholar
- Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39(3):193–206.
- Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100(25):15178–15183.
- Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021.
- Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760.View this article via: PubMed Google Scholar
- Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526.View this article via: PubMed Google Scholar
- Tamura K, et al. Accumulation of CD133-positive glioma cells after high-dose irradiation by Gamma Knife surgery plus external beam radiation. J Neurosurg. 2010;113(2):310–318.
- McCord AM, Jamal M, Williams ES, Camphausen K, Tofilon PJ. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clin Cancer Res. 2009;15(16):5145–5153.
- Sotiropoulou PA, et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol. 2010;12(6):572–582.
- Xi G, et al. CD133 and DNA-PK regulate MDR1 via the PI3K- or Akt-NF-κB pathway in multidrug-resistant glioblastoma cells in vitro. Oncogene. 2016;35(42):5576.
- Zhuang W, Li B, Long L, Chen L, Huang Q, Liang ZQ. Knockdown of the DNA-dependent protein kinase catalytic subunit radiosensitizes glioma-initiating cells by inducing autophagy. Brain Res. 2011;1371:7–15.
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4(9):712–720.
- Diehn M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783.View this article via: PubMed Google Scholar
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284.
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868.
- Bonner WM, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–967.
- Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47(2):320–329.
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72(1):131–142.
- Chan DW, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16(18):2333–2338.
- Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24(6):949–961.
- Lagasse E. Cancer stem cells with genetic instability: the best vehicle with the best engine for cancer. Gene Ther. 2008;15(2):136–142.
- Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–1113.View this article via: PubMed Google Scholar
- Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323.
- Li F, Tiede B, Massagué J, Kang Y. Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res. 2007;17(1):3–14.
- Martín M, et al. Postreplicative joining of DNA double-strand breaks causes genomic instability in DNA-PKcs-deficient mouse embryonic fibroblasts. Cancer Res. 2005;65(22):10223–10232.
- Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 2015;149:124–138.
- Gavande NS, et al. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol Ther. 2016;160:65–83.
- Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2014;40(1):109–117.
- Prevo R, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13(11):1072–1081.
- Batey MA, et al. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol Cancer Ther. 2013;12(6):959–967.
- Biddlestone-Thorpe L, et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin Cancer Res. 2013;19(12):3189–3200.
- Zhao Y, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66(10):5354–5362.
- Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Löbrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64(7):2390–2396.
- Yuan M, Eberhart CG, Kai M. RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget. 2014;5(9):2820–2826.View this article via: PubMed Google Scholar
- Leahy JJ, et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett. 2004;14(24):6083–6087.
- van der Burg M, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119(1):91–98.
- Yan K, et al. Glioma cancer stem cells secrete Gremlin1 to promote their maintenance within the tumor hierarchy. Genes Dev. 2014;28(10):1085–1100.
- Eyler CE, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146(1):53–66.
- Xie Q, et al. RBPJ maintains brain tumor-initiating cells through CDK9-mediated transcriptional elongation. J Clin Invest. 2016;126(7):2757–2772.
- Schonberg DL, et al. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28(4):441–455.
- Xie Q, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18(4):501–510.
- Alan Mitteer R, et al. Proton beam radiation induces DNA damage and cell apoptosis in glioma stem cells through reactive oxygen species. Sci Rep. 2015;5:13961. View this article via: PubMed Google Scholar
- An J, Xu QZ, Sui JL, Bai B, Zhou PK. Downregulation of c-myc protein by siRNA-mediated silencing of DNA-PKcs in HeLa cells. Int J Cancer. 2005;117(4):531–537.
- Olive PL, Wlodek D, Banáth JP. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 1991;51(17):4671–4676.View this article via: PubMed Google Scholar
- Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110.
- Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577(1-2):275–283.
- Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291(5507):1284–1289.
- D’Urso V, Collodoro A, Mattioli E, Giordano A, Bagella L. Cytometry and DNA ploidy: clinical uses and molecular perspective in gastric and lung cancer. J Cell Physiol. 2010;222(3):532–539.View this article via: PubMed Google Scholar
- Pantazi E, et al. GLI2 induces genomic instability in human keratinocytes by inhibiting apoptosis. Cell Death Dis. 2014;5:e1028. View this article via: PubMed Google Scholar
- Iliakis G, Rosidi B, Wang M, Wang H. Plasmid-based assays for DNA end-joining in vitro. Methods Mol Biol. 2006;314:123–131.
- Fenech M, et al. HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res. 2003;534(1-2):65–75.View this article via: PubMed Google Scholar
- Huang M, et al. c-Met-mediated endothelial plasticity drives aberrant vascularization and chemoresistance in glioblastoma. J Clin Invest. 2016;126(5):1801–1814.
- Ciznadija D, Liu Y, Pyonteck SM, Holland EC, Koff A. Cyclin D1 and cdk4 mediate development of neurologically destructive oligodendroglioma. Cancer Res. 2011;71(19):6174–6183.
- Liu Y, et al. Somatic cell type specific gene transfer reveals a tumor-promoting function for p21(Waf1/Cip1). EMBO J. 2007;26(22):4683–4693.
- Fan Y, et al. Profilin-1 phosphorylation directs angiocrine expression and glioblastoma progression through HIF-1α accumulation. Nat Cell Biol. 2014;16(5):445–456.
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111.
-
Version history
- Version 1 (February 8, 2018): Electronic publication











