Research ArticleImmunologyVirology Free access | 10.1172/jci.insight.92424
High-dimensional CyTOF analysis of dengue virus–infected human DCs reveals distinct viral signatures
Rebecca E. Hamlin,1,2 Adeeb Rahman,3,4 Theodore R. Pak,2,4,5 Kevin Maringer,1,6 Ignacio Mena,1 Dabeiba Bernal-Rubio,1 Uma Potla,1 Ana M. Maestre,1 Anthony C. Fredericks,1,2 El-ad D. Amir,3,7 Andrew Kasarskis,2,4,5 Irene Ramos,1 Miriam Merad,2,3,7 and Ana Fernandez-Sesma1,2
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Hamlin, R. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Rahman, A. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Pak, T. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by
Maringer, K.
in:
PubMed
|
Google Scholar
|

1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by
Mena, I.
in:
PubMed
|
Google Scholar
|

1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Bernal-Rubio, D. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Potla, U. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by
Maestre, A.
in:
PubMed
|
Google Scholar
|

1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Fredericks, A. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Amir, E. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Kasarskis, A. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by
Ramos, I.
in:
PubMed
|
Google Scholar
|

1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Merad, M. in: PubMed | Google Scholar
1Department of Microbiology,
2Graduate School of Biomedical Sciences,
3Human Immune Monitoring Core,
4Department of Genetics and Genomic Sciences, and
5Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
6Department of Microbial Sciences, Faculty of Health and Medical Sciences, University of Surrey, Guildford Surrey, United Kingdom.
7Department of Oncological Sciences, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
Find articles by Fernandez-Sesma, A. in: PubMed | Google Scholar
Published July 6, 2017 - More info
JCI Insight. 2017;2(13):e92424. https://doi.org/10.1172/jci.insight.92424.
Copyright © 2017, American Society for Clinical Investigation
Received: December 21, 2016; Accepted: May 19, 2017
-
Abstract
Dengue virus (DENV) is the most prevalent mosquito-borne virus causing human disease. Of the 4 DENV serotypes, epidemiological data suggest that DENV-2 secondary infections are associated with more severe disease than DENV-4 infections. Mass cytometry by time-of-flight (CyTOF) was used to dissect immune changes induced by DENV-2 and DENV-4 in human DCs, the initial targets of primary infections that likely affect infection outcomes. Strikingly, DENV-4 replication peaked earlier and promoted stronger innate immune responses, with increased expression of DC activation and migration markers and increased cytokine production, compared with DENV-2. In addition, infected DCs produced higher levels of inflammatory cytokines compared with bystander DCs, which mainly produced IFN-induced cytokines. These high-dimensional analyses during DENV-2 and DENV-4 infections revealed distinct viral signatures marked by different replication strategies and antiviral innate immune induction in DCs, which may result in different viral fitness, transmission, and pathogenesis.
-
Introduction
Dengue virus (DENV) is the most prevalent mosquito-borne virus causing disease in humans (1), with an estimated 390 million annual infections worldwide, of which 96 million manifest with clinical symptoms (2). Dengue disease may present as a nonspecific febrile illness or as a more severe infection marked by hemorrhage or circulatory failure (3, 4). DENV is a flavivirus that exists as 4 distinct serotypes (DENV-1–DENV-4) and is transmitted by Aedes mosquitoes (5). At the amino acid level, the 4 DENV serotypes share limited similarity among serotypes (about 60%–75%) but about 97% similarity within a given serotype (6). Epidemiological studies have reported differences between serotypes with regard to disease severity. Notably, DENV-2 secondary infections have been associated with more severe disease, while DENV-4 infections have been correlated with more mild disease (7–12).
Hematopoietic cells of the myeloid lineage, such as monocytes, macrophages, and DCs, are the primary targets of DENV infection, replication, and dissemination (13–21). Immature DCs in the periphery efficiently capture antigens via phagocytosis and macropinocytosis. Upon interacting with infectious agents, immature DCs subsequently mature, a process that includes DC surface remodeling, increased antigen presentation, and the production of numerous cytokines (22). Mature DCs upregulate chemokine receptors (e.g., CCR7) that aid in their migration to lymph nodes as well as costimulatory molecules (e.g., CD80, CD86, CD40) that are required for activating antigen-specific T cells in the lymph node and inducing T cell proliferation and effector functions (23, 24). In this way, DCs are key players in linking innate and adaptive immunity. Therefore, the ability of viruses to modulate innate immunity in these cells will have a significant affect on viral replication, dissemination, and pathogenesis.
Peripheral blood mononuclear cells from healthy blood donors can be used to generate monocyte-derived DCs. These cells can be productively infected with DENV and are the primary human ex vivo model for studying primary DENV infections and investigating the effects of DENV infection on DC activation and function (16, 17, 25).
Cytokine production during DENV infection has been observed in patients and in vitro (26). Nevertheless, the pathogenic versus protective roles of cytokines during DENV infection are not completely understood. It has been proposed that a “cytokine storm” results in increased vascular permeability and more severe disease (27). However, there are conflicting reports as to whether the Th1 cytokines IFN-γ and TNF-α as well as the regulatory cytokines IL-10 and TGF-β promote pathogenesis or confer protection during DENV infection (28–30). Additionally, our group and others have demonstrated that DENV-2 (strain 16681) inhibits both type I IFN (IFN-α/β) production and signaling and impairs CD4+ T cell polarization (16, 17, 25, 31–33), which may prevent protective adaptive immune responses.
In this study, we have utilized mass cytometry by time-of-flight (CyTOF) to investigate the effects of DENV infection on human DCs. Unlike flow cytometry, which requires compensation for overlapping fluorescence spectra, CyTOF uses antibodies labeled with elemental isotopes and is largely unencumbered by interference from spectral overlap between channels (34). Currently, CyTOF allows for single-cell analysis for up to 45 parameters simultaneously (35). With this powerful technology, which has not yet been used to characterize infection with DENV or other hemorrhagic fever viruses, we have simultaneously investigated DENV infection, DC activation, intracellular cytokine production, and apoptosis as well as the differences between bystander and infected cells in the same DC culture. Importantly, investigating viral replication and the subsequent host innate immune responses will provide insight into fitness strategies used by vector-borne viruses like DENV to remain in circulation, while inducing varying degrees of disease severity in humans. Herein, we provide a comprehensive functional analysis comparing DC infections with DENV-2 and its most phylogenetically distant serotype, DENV-4, which have been associated with differences in disease severity. We show that DENV-2 and DENV-4 display different replication strategies and induce different antiviral innate immune responses in human DCs, which may be related to differences in viral fitness, transmission, and pathogenesis.
-
Results
Replication differences between DENV-2 and DENV-4. In this study, we infected human donor-matched DCs with DENV-2 and DENV-4 (MOI of 0.5). The viral isolates used were representative of prevalent genotypes of the DENV-2 and DENV-4 serotypes (Supplemental Figures 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.92424DS1). Using CyTOF analysis to measure intracellular DENV structural and nonstructural proteins and titration of extracellular infectious DENV particles, we found that DENV-2 peak infection occurred at 48 hours after infection (hpi), whereas DENV-4 infection peaked at 24 hpi and was significantly diminished by 72 hpi (Figure 1, A–D). Notably, DENV-4 infection resulted in a greater percentage of infected DCs compared with DENV-2 infection (Figure 1, A and B). Interestingly, despite significant differences in the percentages of infected cells (Figure 1B) and different infection kinetics (Figure 1D), the peak titers of infectious particles released extracellularly were similar between DENV-2 and DENV-4 (Figure 1D). Additionally, even though DENV-2 infected a smaller percentage of cells compared with DENV-4, those DCs infected with DENV-2 showed significantly higher levels of DENV nonstructural 3 (NS3) protein (Figure 1C) at 48 and 72 hpi compared with the DCs infected with DENV-4. Together, these data suggest that DENV-2 infects fewer DCs later during infection, with each infected cell producing many infectious particles, whereas DENV-4 infects more DCs earlier during infection, with each infected cell producing fewer infectious particles.
 Figure 1
Figure 1DENV-2 and DENV-4 demonstrate replication differences in human DCs but not in mosquito cells. (A) Mass cytometry by time-of-flight (CyTOF) analysis of DCs infected at an MOI of 0.5 with dengue virus 2 (DENV-2) and DENV-4. Viral infection was visualized by antibody staining for the DENV envelope (DENV E) and nonstructural 3 (NS3) proteins. DENV-infected cells were gated on live single CD45+ cells. One representative donor of seven is shown. (B) The percentage of DENV-infected DCs was quantified as shown in A by CyTOF analysis for all 7 donors. (C) Mean signal intensity of DENV NS3 protein staining for cells positive for DENV infection for all 7 donors. (D) Replication kinetics of DENV-2 and DENV-4 in DCs, as determined by plaque assay of extracellular infectious particles. Fifteen donors are represented. (E) Replication kinetics of DENV-2 and DENV-4 (MOI of 0.5) in Aag2 cells, as determined by plaque assay of extracellular infectious particles. The graph depicts 3 biological replicates. Mean ± SEM is shown. Black asterisks represent statistical significance by the paired, 2-tailed Wilcoxon signed-rank test, comparing DENV-4 to DENV-2 at each time point. Red asterisks represent statistical significance for either DENV-2 or DENV-4 compared with mock-infected cells at each time point. The Benjamini-Hochberg procedure was performed within each time point to adjust the significance level for multiple comparisons (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
We also compared DENV-2 and DENV-4 replication kinetics in mosquito cells. Using Aag2 cells (an Aedes aegypti cell line), we observed comparable replication kinetics for DENV-2 and DENV-4 during a 5-day time course (Figure 1E). In particular, the rapid peak and subsequent decline in titers for DENV-4 during DC infection was not observed in mosquito cells. Thus, these viruses have different replication kinetics, and presumably different viral fitness strategies, in human DCs but not in mosquito cells.
Differential effects of DENV-2 and DENV-4 infection on DC phenotype and function. To fully appreciate DC phenotype and function during DENV infection, three distinct yet complementary techniques were employed to analyze the CyTOF data: (a) a global view of the DC phenotype was demonstrated by heat maps utilizing an autogating strategy (36, 37) (Figure 2 and Supplemental Figure 3B); (b) multiple DC donors were compared via a manual gating scheme (Supplemental Figure 3A and Supplemental Figures 4–6); and (c) single-cell data were illustrated with viSNE analysis (Figure 3 and Supplemental Figure 7). With these analyses, it was apparent that DC infection with DENV-2 or DENV-4 led to significant upregulation of DC activation markers CD80 and CD40 as well as cytokines and chemokines (i.e., TNF-α, IL-1β, IL-8, and IP-10) compared with mock treatment (Figure 2, A and B; Figure 3; and Supplemental Figures 4, 5, and 7). Additionally, DENV-4 infection, but not DENV-2 infection, led to the upregulation of DC migration marker CCR7 compared with mock treatment (Figure 2, A and B; Figure 3; and Supplemental Figures 4 and 7). Strikingly, we found that DENV-4 infection induced higher expression of CD80, CD40, and CCR7 and greater production of TNF-α+, IL-1β+, and IL-8+ cells when compared with DENV-2 infection (Figure 2C; Figure 3; and Supplemental Figures 4, 5, and 7). Notably, DCs infected with DENV-2 and DENV-4 did not demonstrate statistically significant differences in expression of cleaved caspase-3, a marker for apoptosis, or in levels of cell death (Supplemental Figure 6) (38).
 Figure 2
Figure 2Comparing cellular marker positivity during mock, DENV-2, and DENV-4 infections by autogating. Heat maps comparing cellular marker positivity by denoting cells as either positive or negative for each marker during mock, dengue virus 2 (DENV-2), and DENV-4 infections. For all plots, markers are ordered vertically by hierarchical clustering of the rows, which places markers with similar expression profiles closer together. Statistical significance was determined using a 2-sided Fisher’s exact test at a threshold of α = 10–5 with Bonferroni correction for multiple testing. Cellular markers not representing a significant difference between infection conditions at this threshold are labeled with “NS”; all others are significant. Calculations were made using the event counts from mass cytometry by time-of-flight (CyTOF) data, as determined by autogating of cells positive for each marker based on the 99th percentile threshold, which was set for mock treatment at 8 hours after infection (hpi). The comparative effects on each marker were calculated as the log10 of the relative risk for positive marker frequency between the indicated infection conditions. The histograms within the heat map color scales show the total number of cells matching that intensity range, revealing the overall distribution of the values. One representative DC donor of seven is shown. (A) DENV-2 infection and mock treatment are compared. Red indicates greater marker expression during DENV-2 infection, and blue indicates greater marker expression during mock treatment. (B) DENV-4 infection and mock treatment are compared. Red indicates greater marker expression during DENV-4 infection, and blue indicates greater marker expression during mock treatment. (C) DENV-4 and DENV-2 infections are compared. Red indicates greater marker expression during DENV-4 infection, and blue indicates greater marker expression during DENV-2 infection. White indicates no difference in marker expression between the infection conditions.
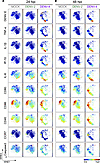 Figure 3
Figure 3The phenotypic landscape is different during DENV-2 versus DENV-4 infection in DCs. Live single CD45+ cells from representative donor 2 (Supplemental Figure 3B) of the 7 DC donors were used to create viSNE plots, which were generated using all 18 phenotypic markers used for mass cytometry by time-of-flight (CyTOF) staining except markers for dengue virus (DENV). Each point in the plot represents a single cell, and the axes show arbitrary units based on the t-distributed stochastic neighbor embedding (t-SNE) algorithm. The relative position of a cell on the plot represents its general phenotype in multidimensional space. For (A) 24 hours after infection (hpi) and (B) 48 hpi, cells are colored according to staining intensity of the indicated marker, enabling the comparison of expression patterns across markers for different infection conditions and time points after infection.
Additionally, with viSNE analysis of DENV-4 infection, it was notable that there were clusters of cells expressing high levels of DENV envelope (DENV E) protein, but these cells were not limited to one phenotypic DC population (Figure 3 and Supplemental Figure 7B). Interestingly, cells expressing high levels of TNF-α and IL-1β corresponded with a cluster of DENV E–expressing cells (Figure 3 and Supplemental Figure 7B). IL-8 expression was more broadly distributed but also largely colocalized to the same region of the viSNE plots (Figure 3 and Supplemental Figure 7B). In contrast, cells most highly expressing IP-10 did not correspond with DENV E–expressing cells (Figure 3). During the peak of DENV-2 infection, a small number of cells expressing high levels of DENV E were visible but distributed throughout the viSNE plots (Figure 3 and Supplemental Figure 7B). In addition, TNF-α, IL-1β, IP-10, and IL-8 were notably upregulated in DCs during DENV-2 infection compared with mock treatment (Figure 3 and Supplemental Figure 7).
Interestingly, DC costimulatory markers CD80, CD86, and CD40 as well as the DC migration marker CCR7 displayed an entirely different pattern, with cells expressing high levels of these markers being localized to distinct regions of the plots from those expressing high levels of cytokines; this was most clear during later time points of DENV-4 infection (Figure 3 and Supplemental Figure 7B). This indicates that the cells upregulating cytokines are distinct from those upregulating high levels of DC maturation markers during DENV infection. This was consistent with the hierarchical clustering analysis of autogated CyTOF data (Figure 2), in which cytokines clustered more closely to DENV antigens and other cytokines and farther from DC phenotypic and functional markers. Additionally in viSNE analysis, the cells expressing high levels of cleaved caspase-3, a marker for apoptosis, were largely localized to a distinct cluster, representing another distinct group of cells responding to DENV infection (Figure 3 and Supplemental Figure 7). Altogether, these data indicated that the responses to DENV infection in these DC cell cultures were heterogeneous and that there were distinct cellular phenotypes associated with cells expressing viral antigens.
The contribution of bystander versus infected cells during DENV infection. The previous data (Figures 2 and 3) revealed marked phenotypic differences induced by DENV-2 and DENV-4 for the entire population of each DC sample. The power of CyTOF analysis includes the possibility of distinguishing between infected and bystander cells within a sample. This allowed us to interrogate the specific contribution of bystander versus DENV-infected cells by using markers indicative of DENV infection (i.e., DENV E and NS3 proteins). Notably, the various patterns observed between infected and bystander cells were similar during both DENV-2 and DENV-4 infections. Generally, DC activation markers CD80, CD86, CD40, and CCR7 were similarly upregulated by DENV-infected and bystander cells following both DENV-2 and DENV-4 infection of DC cultures (Figure 4). On the other hand, both DENV-2–infected and DENV-4–infected DCs showed increased production of TNF-α+ and IL-1β+ cells compared with their respective bystander cells (Figure 5, A and B). Remarkably, this was notable even for DENV-2, despite the low number of DENV-2–infected cells. IL-8 production was generally similar between DENV-infected and bystander cells, except at an early time point when DENV-4–infected cells demonstrated a greater percentage of IL-8+ cells compared with their bystander cells (Figure 5C). Interestingly, and consistent with its role as an IFN-stimulated gene (ISG), IP-10 was produced primarily by bystander cells compared with DENV-4–infected cells at 24 and 48 hpi, but this effect was not as pronounced in DENV-2–infected cultures (Figure 5D). Thus, proinflammatory cytokines (TNF-α and IL-1β), chemokines (IL-8), and ISGs (IP-10) demonstrated distinct patterns of production in DENV-infected versus bystander DCs. Additionally, both DENV-2– and DENV-4–infected DCs demonstrated higher percentages of cleaved caspase-3+ cells compared with their respective bystander cells at later time points, suggesting greater levels of apoptosis in DENV-infected cells (Figure 5E). It is notable that the timing of these observations is consistent with the kinetics of infection for these two viruses.
 Figure 4
Figure 4Bystander cells show similar DC surface marker phenotypes compared with infected cells during DENV infection. Dengue virus–infected (DENV-infected) DCs were gated on live single CD45+ cells, as shown in Supplemental Figure 3A. Using mass cytometry by time-of-flight (CyTOF) analysis for each DENV-2 or DENV-4 sample, the cells were stratified as DENV-infected (I) cells (filled circles) or bystander (B) cells (open circles). Mock-infected cells (M) were analyzed as a single population. Seven DC donors are shown. Median expression levels of DC surface markers (A) CD80, (B) CD86, (C) CD40, and (D) CCR7 for mock, bystander, and DENV-infected cells of DENV-2 (green) and DENV-4 (purple) samples are shown. Mean ± SEM is shown. Black asterisks represent statistical significance by the paired, 2-tailed Wilcoxon signed-rank test, comparing bystander and infected cells within a given culture sample at a given time point. The Benjamini-Hochberg procedure was performed within each time point to adjust the significance level for multiple comparisons (*P ≤ 0.05).
 Figure 5
Figure 5Bystander cells show distinct cytokine and cleaved caspase-3 phenotypes compared with infected cells during DENV infection. Dengue virus–infected (DENV-infected) DCs were gated on live single CD45+ cells, as shown in Supplemental Figure 3A. Using mass cytometry by time-of-flight (CyTOF) analysis for each DENV-2 or DENV-4 sample, the cells were stratified as DENV-infected (I) cells (filled circles) or bystander (B) cells (open circles). Mock-infected cells (M) were analyzed as a single population. Seven DC donors are shown. The percentage of mock, bystander, or DENV-infected cells of DENV-2 (green) and DENV-4 (purple) samples that stained positive intracellularly for the cytokines (A) TNF-α, (B) IL-1β, (C) IL-8, and (D) IP-10 and for the apoptosis marker (E) cleaved caspase-3 are shown. Mean ± SEM is shown. Black asterisks represent statistical significance by the paired, 2-tailed Wilcoxon signed-rank test, comparing bystander and infected cells within a given culture sample at a given time point. The Benjamini-Hochberg procedure was performed within each time point to adjust the significance level for multiple comparisons (*P ≤ 0.05).
Cytokine secretion during DENV-2 versus DENV-4 infection. In addition to intracellular cytokine analyses by CyTOF, multiplex ELISAs were performed to evaluate extracellular cytokine secretion by DCs during DENV infection. Consistent with prior results (16, 17, 25, 31, 32, 39, 40), DENV-2 infection induced minimal IFN-α and low levels of IP-10 secretion (Figure 6). In contrast, DENV-4 infection of DCs induced greater IFN-α and IP-10 secretion compared with mock infection and DENV-2 infection of DCs (Figure 6). Infection with DENV-4 also induced greater secretion of several proinflammatory cytokines and chemokines (i.e., TNF-α, IL-6, IL-1β, IL-12, MIP-1α, MIP-1β, RANTES, and IL-8) compared with mock and DENV-2 infection (Figure 6). Of note, DENV-2 infection induced the secretion of several proinflammatory cytokines and chemokines compared with mock infection but to a lesser extent than DENV-4 infection (Figure 6).
 Figure 6
Figure 6DENV-4 infection of DCs induces greater cytokine and chemokine secretion compared with DENV-2 infection. Levels of cytokine and chemokine secretion (as indicated) were measured by performing multiplex ELISA on supernatants of DC cultures infected with dengue virus 2 (DENV-2) or DENV-4 (MOI of 0.5) or mock-infected cultures. Sixteen donors are shown. Mean ± SEM is shown. Black asterisks represent statistical significance by the paired, 2-tailed Wilcoxon signed-rank test, comparing DENV-4 to DENV-2 at each time point. Red asterisks represent statistical significance for either DENV-2 or DENV-4 compared with mock-infected cells at each time point. The Benjamini-Hochberg procedure was performed within each time point to adjust the significance level for multiple comparisons (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
The continuous upregulation of cytokine and chemokine production during DENV-2 and DENV-4 infection appears to be replication dependent, since infection of DCs with either UV-inactivated DENV-2 or UV-inactivated DENV-4, which did not produce infectious particles, resulted in lower levels of cytokine and chemokine secretion (e.g., IL-6, TNF-α, RANTES, IP-10, MIP-1α, and MIG) at later time points compared with infection with their respective, replication-competent DENV (Supplemental Figure 8).
The observed differences in cytokine and chemokine production by DENV-2 and DENV-4 could be theoretically due to the different proportion of infected cells in the culture between the two viruses. To address this possibility, we infected DCs with DENV-2 and DENV-4 at different MOIs to yield a similar percentage of infected cells at their respective peak titers (Supplemental Figure 9). We found that cytokine and chemokine secretion for DENV-2 infection was generally MOI dependent, with greater cytokine and chemokine secretion at the higher MOI (Supplemental Figure 9). Nevertheless, even when DCs were infected with DENV-2 at a high MOI of 25, cytokines and chemokines (e.g., IL-6, TNF-α, IL-1β, IL-12, RANTES, IP-10, MIP-1α, MIP-1β, and MIG) were still secreted at higher levels during infection with DENV-4 (MOI of 0.5) at most time points (Supplemental Figure 9). Together, these data suggest that the level of cytokine and chemokine secretion depended less on viral load (MOI) and more on inherent strain differences between the two virus strains.
-
Discussion
Human immune responses induced by infection with DENV and other hemorrhagic fever viruses are complex and characterized by high levels of proinflammatory cytokines and chemokines. CyTOF allows for the simultaneous examination of viral infection, DC activation, cytokine production, and apoptosis for individual cells. In this work, we provide a comprehensive, detailed analysis of the infection kinetics and immune responses induced by DENV-2 and DENV-4 strains in human DCs, which revealed distinct viral signatures. To our knowledge, this is the first study to utilize CyTOF to examine cells infected with a hemorrhagic fever virus.
Interestingly, these DENV-2 and DENV-4 strains show markedly different replication kinetics in human DCs but replicate similarly in mosquito cells (Figure 1). This DENV-4 strain is also more capable of activating human DCs, upregulating DC migration markers, and inducing cytokine production compared with this DENV-2 strain. Therefore, we postulate that DENV-4 may be better adapted for prolonged virus replication in mosquitoes compared with human cells, whereas DENV-2 may be well adapted for prolonged replication in both hosts. The early burst in DENV-4 replication may be sufficient to produce the necessary level of viremia in the human host to allow for early transmission back to the mosquito before the human immune response effectively clears the infection, thereby preventing continuous inflammatory responses caused by sustained viral replication. This viral fitness strategy could enable DENV-4 to continue circulating without causing severe disease in most infected humans. In contrast, the DENV-2 strain infects relatively few cells and does not induce such a robust innate immune response in human DCs. In this way, DENV-2 may be more efficient than DENV-4 at evading human immune responses, causing a more prolonged viremia in the human host and ensuring later transmission back to the mosquito. Additionally, the limited induction of innate immunity during DENV-2 infection may lead to inefficient adaptive immune responses (33) and greater epidemic potential for DENV-2. The data presented in this work thereby support the epidemiological studies showing that DENV-4 infections generally result in milder disease in humans compared with DENV-2 (7–12). Such pronounced differences in immune responses induced by these DENV serotypes illustrate the importance of continued investigation into the unique qualities of each DENV serotype to better characterize DENV-induced immune responses and pathogenesis. Furthermore, since the representative DENV strains used in this study were isolated many years ago and have therefore undergone numerous passages in the laboratory, future studies will be important for testing multiple strains of these DENV serotypes, including contemporary strains and low-passage human isolates, to further test these hypotheses and to minimize the effects of possible viral adaptations that can occur during passage in tissue culture. Additionally, future studies will investigate blood samples from dengue-infected patients to correlate virus replication and immune profiles with DENV serotype and disease severity.
There are many differences between these two DENV serotypes that may account for the observed differences in replication and immune induction in human cells, which can be tested in future studies. First, it has been shown that DENV-4 NS5 is predominantly located in the cytoplasm, whereas DENV-2 NS5 is predominantly located in the nuclei of infected cells (41–43). Since DENV replication takes place in the cytoplasm (44), there may be a correlation between the availability of more NS5 polymerase molecules for DENV-4 replication in the cytoplasm and earlier peak replication kinetics. In addition, differences in DENV NS5 nuclear localization may affect splicing of host RNAs, such as those involved in immune responses to virus infections (45). Additionally, the most 5′ region of the DENV 3′ untranslated region is one of the least conserved regions of the DENV genome among the serotypes (46). Due to differences in this region, DENV-2 and DENV-4 likely produce different subgenomic flavivirus RNAs (sfRNAs). It has been proposed that sfRNA accumulation is associated with viral epidemiological fitness (46, 47). In particular, DENV-2 strains with greater sfRNA production relative to viral genomic RNA have been correlated with epidemiological fitness (47). It has also been shown that sfRNA can antagonize human innate immune pathways, decreasing both IFN and ISG production (48). Furthermore, it was recently found that different DENV sfRNA species accumulate in vertebrate and invertebrate hosts during host change, with subsequent implications for innate immune responses and viral fitness (49). It would therefore be very interesting to determine if differences in sfRNA could account for differences between DENV-2 and DENV-4 with regard to innate immune induction and viral fitness during host adaptation between humans and mosquitoes.
Additionally, the viSNE analysis in this study is elucidating because it demonstrates that the DCs producing the highest amount of cytokines are largely distinct from those upregulating the highest amount of DC activation markers during DENV infection (Figure 3 and Supplemental Figure 7B). This is consistent with hierarchical clustering analysis of autogated CyTOF data, in which cytokines tend to cluster more closely with other cytokines and to DENV antigens and in distinct branches from the DC phenotypic and functional markers (Figure 2). This suggests that the expression of different cytokines may be driven by signaling pathways and transcriptional programs that are more similar to one another and less similar to those regulating DC surface marker expression following DENV infection.
In addition to comparing the overall effects of infection by these DENV-2 and DENV-4 strains, we also stratified the CyTOF data to quantitatively compare the effects of DENV on infected cells and bystander cells in the same DC cultures. Existing studies have shown some differential effects of DENV-2 infection on bystander cells versus infected cells in the same culture (50–56). Our unique CyTOF study using 20 markers simultaneously provides more comprehensive data on changes to DC phenotype and function, as well as better temporal resolution, following both DENV-2 and DENV-4 infections. Consistent with prior data (51, 56), we observed that DCs infected with both DENV-2 and DENV-4 strains preferentially produce TNF-α and IL-1β, while bystander DCs preferentially produce IP-10 (Figure 5). Interestingly, our data also show that type I IFN is secreted at higher levels during DENV-4 infection of DCs as compared with DENV-2 infection of DCs (Figure 6). Thus, it is likely that DENV-4–infected cells produce type I IFN, which then induces the production of ISGs, such as IP-10, in bystander cells. These bystander cells should be unable to block IFN signaling, since they do not contain DENV NS5 protein, which degrades STAT2 in infected cells (31). These data are also in agreement with the literature showing that DENV-2 infections do not induce robust type I IFN responses in human DCs (51, 56).
On the other hand, we observed that both DENV-2–infected and DENV-4–infected cells and their respective bystander cells upregulate costimulatory molecules compared with mock-infected DCs, which is consistent with some prior work with DENV-2 (51, 56). The upregulation in CCR7 observed in our study for both infected and bystander cells after DENV-4 infection suggests that both DENV-4–infected and bystander DCs could migrate to draining lymph nodes during an in vivo infection. Since we observed that both infected and bystander DCs are capable of activation, the bystander DCs are most likely undergoing cytokine-mediated maturation (51, 52). Of note, it has been shown that cytokine-induced maturation of DCs and subsequent neoantigen pulsing promotes DC priming of naive CD4+ T cells toward a Th1 response (23).
Furthermore, we found that DENV-infected cells preferentially undergo apoptosis (as determined by cleaved caspase-3 expression) compared with bystander cells at times consistent with viral replication during both DENV-2 and DENV-4 infections (Figure 5E). This is consistent with other work showing that sustained DENV infection may lead to greater induction of apoptosis (55). Cells undergoing apoptosis may release viral progeny, contributing to virus spreading, or may release apoptotic bodies that are later engulfed by other DCs, which may lead to bystander DC maturation. It is also possible that loss of DENV-infected DCs by apoptosis could impair the connection between innate and adaptive immunity and thereby diminish the human immune response to DENV infection. With these data, we have demonstrated that both sample-wide and population-stratified analyses are valuable and yield different insights into the immunological consequences of viral infections. Our data clearly demonstrate that DENV infection induces important phenotypic changes in both infected and bystander cells, both of which likely contribute to the overall immune response to these viral infections. Furthermore, these data illustrate that even small numbers of DENV-infected cells can contribute to these different responses.
This work, in concert with previous data in the literature, suggests the following model: dermal DCs and Langerhans cells are the primary cells infected with DENV following the bite of an infected mosquito (21, 57–59). The infected DCs secrete proinflammatory cytokines and chemokines, resulting in an influx of monocytes into the dermis that can differentiate into DCs and become new targets for DENV replication (59). Infected DCs undergo activation and maturation, which is characterized by the upregulation of markers, such as CD40, CD80, CD86 and CCR7. Cytokine production (e.g., TNF-α and IL-1β) by DENV-infected cells can activate bystander DCs, causing them to mature and upregulate CCR7. Other immune stimuli, such as defective viral particles or DENV-infected DC-derived exosomes containing viral components (60–64), could also activate bystander cells. Both infected and bystander DCs are then able to migrate to draining lymph nodes and interact with T cells (57). Bystander cells could also have been abortively infected or could have captured DENV antigens via phagocytosis of exosomes containing DENV antigens; those antigens would have been missed with the antibody used to measure active replication in our assay. Thus, it is possible that bystander cells may still be able to present DENV antigens to T cells in the lymph nodes. On the other hand, it is also possible that bystander cells may not have acquired DENV antigens, and their migration to the draining lymph nodes may interfere with T cell priming by DENV-infected DCs. The magnitude of type I IFN and IP-10 production by DCs may also contribute to the ability of these DCs to prime T cells in the lymph node toward a Th1 response (16). In this way, infection of DCs with this DENV-4 strain, which results in greater production of IFN-α, IP-10, and IL-12, may be more efficient at skewing the adaptive immune response toward a Th1 response compared with infection with this DENV-2 strain.
Altogether, this study illustrates the power of utilizing CyTOF to examine the complex immune phenotypes following virus infection. Indeed, using this technology, we provide insights into the differences between serotypes and bystander cells versus infected cells. From our data, we also postulate that different DENV strains use different replication strategies to produce a sufficiently high viremia for transmission back to the mosquito vector; these differences in replication will also greatly affect innate immune induction in the host. Finally, this work may provide a useful framework for investigating emerging viruses, in particular, arboviruses, like chikungunya virus and Zika virus, that have caused recent epidemics. Investigating different replication strategies used by vector-borne viruses and the consequential immune induction by the host may provide insights into different fitness strategies that vector-borne viruses use to remain in circulation.
-
Methods
Data is available at http://immport.org from study accessions SDY1100 and SDY1149.
Cell lines. Baby hamster kidney (BHK) cells (originally obtained from Sujan Shresta, La Jolla Institute for Allergy and Immunology, La Jolla, California, USA) were cultured at 37°C in α-MEM GlutaMAX, supplemented with 10% (v/v) FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 mM HEPES. These tissue culture reagents were purchased from Thermo Fisher Scientific. Aag2 (Aedes aegypti) cells were a gift from Raul Andino (University of California, San Francisco, San Francisco, California, USA) and were maintained in Leibovitz’s L-15 media (Thermo Fisher Scientific) supplemented with 8% (v/v) tryptose phosphate broth, 2 mM L-glutamine, 0.1 mM MEM nonessential amino acids (Sigma-Aldrich), 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% (v/v) FBS (all Thermo Fisher Scientific) at 28°C in a humidified atmosphere without CO2. C6/36 (Aedes albopictus) cells (originally obtained from Jorge Munoz-Jordan, Centers for Disease Control and Prevention, San Juan, Puerto Rico) were maintained in RPMI media (Invitrogen) supplemented with 0.15% (m/v) sodium bicarbonate (Sigma-Aldrich), 1X MEM nonessential amino acids, 2 mM L-glutamine, 1 mM sodium pyruvate, and 10% (v/v) FBS (all from Thermo Fisher Scientific) at a 33°C (with 5% CO2) incubator.
All cell lines used herein were tested for the presence of Mycoplasma using the Mycoalert mycoplasma detection kit from Lonza (catalog LT07-118). Cells and virus preparations used in these experiments were all Mycoplasma free.
Virus preparations. A prototypic DENV-2 strain belonging to the Asian I genotype (Supplemental Figure 2) that was isolated from a patient with dengue shock syndrome in Thailand in 1964 (DENV-2 16681) (65, 66) and a DENV-4 strain belonging to genotype IIB (Supplemental Figure 1) that was isolated from a patient with dengue fever in Indonesia in 1976 (DENV-4 1036) (67) were used in this study (68). DENV-2 16681 (cDNA) and DENV-4 1036 were initially obtained from Richard Kinney (Arbovirus Disease Branch, Centers for Disease Control and Prevention, Fort Collins, Colorado, USA) and underwent minimal subsequent passages in C6/36 cells in our laboratory. These viruses were both grown in C6/36 insect cells for 7 days as described elsewhere (69). In brief, C6/36 cells were infected with either DENV-2 or DENV-4 at MOI of 0.01; 7 days after infection, cell supernatant was collected and stored at –80°C. DENV stock titers were determined by limiting-dilution plaque assays on BHK cells as described below. UV-inactivated DENV-2 and DENV-4 were prepared by irradiating the virus with a UV lamp for 10 minutes (at 6 inches); virus inactivation was confirmed by plaque assay on BHK cells.
The complete genome sequence of the DENV-4 isolate was obtained by sequencing overlapping RT-PCR products (1.0- to 1.5-Kbp length) using viral RNA purified from DCs infected with DENV-4 as template. A set of 10 primer pairs was designed based on the sequence of available DENV-4 complete genomes. Each primer included the sequence of universal primers M13-fwd (GTAAAACGACGGCCAGT, forward primers) or M13-rev (CAGGAAACAGCTATGAC, reverse primers) to facilitate sequencing. High-fidelity RT-PCR was performed using the Superscript III high-fidelity RT-PCR kit (Invitrogen) and the conditions recommended by the manufacturer. RT-PCR products were purified from an agarose gel and sequenced using universal primers M13-fwd and M13-rev. Finally, the complete genome was manually assembled from the overlapping sequences and has been uploaded to GenBank with accession number KX812530 (https://www.ncbi.nlm.nih.gov/nuccore/KX812530).
To construct a phylogenetic tree, a selection of complete genomes of DENV-4 was downloaded from the NIAID Virus Pathogen Database and Analysis Resource (70) through the website (http://www.viprbrc.org/). The selection was manually trimmed to contain 57 isolates representative of the complete genetic diversity, and the sequence of Indonesia 1976 (strain 1036) was included. A consensus neighbor-joining phylogenetic tree from 1,000 replicates was obtained using the program Geneious (http://www.geneious.com/). The tree was visualized using FigTree (Supplemental Figure 1) (available at http://tree.bio.ed.ac.uk/software/figtree/) and rooted at midpoint for clarity. The genomes used in the alignment were as follows: Brazil2-2010: JN983813.1; Brazil3-2010: JN559741.2; Venezuela5-2007: HQ332176.1; Venezuela4-2007: GQ868644.1; Colombia-2005: GQ868585.1; Venezuela3-2007: HQ332175.1; Venezuela-2007: FJ182017.1; Venezuela2-2007: FJ882582.1; Brazil-2010: JQ513333.1; Colombia-2004: GQ868584.1; Venezuela-2000: FJ850095.1; Colombia-2001: GQ868579.1; Venezuela-1998: FJ639739.1; USA2-1998: EU854296.1;USA-1986: EU854295.1; USA-1999: FJ882599.1; USA2-1996: GQ199882.1; USA-1998: FJ882596.1; USA2-1994: GQ199878.1; USA-1996: GQ199881.1; Haiti-2014: KT276273.1; Haiti-1994: JF262782.1; USA-1994: GQ199879.1; Dengue_814669: AF326573.1; Senegal-1953: KF907503.1; Colombia-1982: GU289913.1; Brazil-1976: JN559740.2; Venezuela-1995: JF262781.1; Indonesia-1976: KX812530; Singapore-1995: AY762085.1; New-Caledonia-2009: JQ915089.1; French-Polynesia-2009: JQ915082.1; French-Polynesia-2010: JQ915084.1; Indonesia2-2007: KC762697.1; Thailand-2000: AY618993.1; China-2010: KP723482.1; Indonesia-2007: KC762696.1; New-Caledonia-2008: JQ915085.1; Singapore-2010: JX024758.1; Singapore-2005: GQ398256.1; China-2012: KC333651.1; China-1978: FJ196849.1; Cambodia-2008: JN638570.1; Thailand-2001: AY618992.1; Thailand-1991: AY618990.1; Cambodia-2002: KF955510.1; Thailand-1977: AY618991.1; Pakistan-2009: KF041260.1; India-2009: JQ922560.1; India-1979: JQ922559.1; South-Korea: KP406806.1; Philippines-1956: GQ868594.1; China: AF289029.1; China-1990: FJ196850.1; India-1962: JQ922558.1; India-1961: JF262783.1; Thailand-1997: AY618988.1; MONKEY-Malaysia-1975: JF262779.1.
Similarly, a consensus neighbor-joining phylogenetic tree of 55 DENV-2 complete genomes was made (Supplemental Figure 2). Genomes used in the alignment were as follows: VietNam2/2006: EU482654; VietNam-N/A: JQ045677; Thailand/2001: FJ639832; Thailand/1988: DQ181802; N/A-1996: AF100463; Thailand/1993: AF022439; Thailand/1994: EU726767; Thailand/1979: DQ181805; Thailand/1984: DQ181804; Thailand/1974: GU289914; Thailand/1964: GQ868591; DENV-2 16681-Thailand/1964: U87411.1; USA/2007: JF730050; USA/2010: HQ541799; Colombia/1944: EU854293; Cuba/1981: KF704354; Indonesia/1975: GQ398268; Philippines/1996: KF744404; USA/2000: EU687222; Puerto Rico/2006: KF955364; USA/1996: EU596486; Venezuela/1991: GQ868596; Puerto Rico/1986: KF955363; USA/1989: EU482580; Mexico/2010: KJ189309; Thailand/1990: DQ181801; VietNam3/2006: EU482672; China/2010: JX470186; Singapore/2006: JN851128; Singapore/2010: KM279579; Taiwan/2002: DQ645553; India/2006: FJ898454; Pakistan/2010: KF360005; India/1996: JQ922549; N/A-1999: AF359579; Fiji/1971: HM582101; New Caledonia/1972: HM582103; French Polynesia/1973: HM582110; Tonga/1974: HM582111; Puerto Rico/1969: JX966380; Puerto Rico/1977: EU056812; Mexico/1992: GQ868590; Mexico/1994: JX966379; Mexico/1983: GQ868588; Colombia/1986: GQ868592; India/1971: JQ922550; India/1980: JQ922553; India/1960: JQ922552; Burkina Faso/1980: EF105386; Cote d’Ivoire/1980: EF105381; Senegal/1999: EF105390; Senegal/1970: EF105384; Senegal/1974: EF105385; Nigeria/1966: EF105388; Malaysia/1970: EF105379.
DENV titration by plaque assay. DENV infectious particles were titrated by plaque assay using BHK cells and a standard protocol (65). BHK cells were seeded in 12-well plates (at 1 × 105 cells/well) and incubated overnight at 37°C with 5% CO2. The following day, the medium was removed and serial dilutions of virus supernatants in α-MEM GlutaMAX were added to the wells (150 μl virus inoculum/well). The cells were then incubated at 33°C and 5% CO2 for 2 hours. Following this incubation, an overlay consisting of 1% low-melting-point Lonza SeaPlaque Agarose in distilled H2O (Thermo Fisher Scientific), 5% FBS, and 45% 2X MEM (Thermo Fisher Scientific) was added at 1.5 ml/well, and the plates were incubated at 33°C with 5% CO2 for 7 days. Subsequently, after removing the overlay, the plaques were fixed and visualized with a 1% crystal violet solution in 20% ethanol for >10 minutes.
Generation of monocyte-derived DCs. Monocyte-derived DCs were generated from buffy coats of healthy human donors (New York Blood Center) using a standard protocol (16, 17, 25, 71). Briefly, peripheral blood mononuclear cells were isolated using Ficoll gradient centrifugation (Histopaque, Sigma-Aldrich). Next, CD14+ cells were isolated from the mononuclear fraction using the MACS CD14+ isolation kit (Miltenyi Biotech) according to the manufacturer’s instructions. CD14+ cells were then differentiated into naive immature DCs by incubation in DC media (RPMI medium with 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1 mM sodium pyruvate [Thermo Fisher Scientific]) supplemented with 500 U/ml human granulocyte-macrophage colony-stimulated factor (GM-CSF) (PeproTech), 1,000 U/ml human IL-4 (PeproTech), and 10% (v/v) Hyclone FBS (Thermo Fisher Scientific) for 5 days at 37°C and 5% CO2.
DENV infection of DCs. After 5 days in culture, donor-matched naive DCs were infected with DENV-2 or DENV-4 at an MOI of 0.5 or treated with UV-inactivated virus or DC medium (mock) for 60 minutes at 37°C with 5% CO2. After virus adsorption, the virus inoculum was removed following centrifugation at 400 g for 10 minutes, and the DCs were then maintained in culture at 1 × 106 cells/ml in DC media containing 10% FBS at 37°C and 5% CO2 for different time periods, depending on the experiment. For infections with UV-inactivated DENV, the same volume of UV-inactivated DENV was added compared with infectious DENV.
DENV infection of Aag2 cells. Subconfluent Aag2 cells cultured in 24-well plates were washed once in PBS 24 hours after seeding and then infected with DENV at an MOI of 0.5 in 200 μl PBS. Following a 1-hour incubation at 28°C, the inoculum was removed and cells were washed once in PBS and then returned in Aag2 culture media to the 28°C incubator. Supernatants were harvested at indicated time points and titrated on BHK cells as previously described.
Multiplex ELISA. The Cytokine Human Magnetic 25-Plex Panel for Luminex Platform (Thermo Fisher Scientific) was used according to the manufacturer’s instructions. This kit can detect the following cytokines and chemokines: GM-CSF, TNF-α, IL-1β, IL-4, IL-6, MIP-1β, Eotaxin, RANTES, MIG, IL-12 (p40/p70), IL-8, IL-17, MIP-1α, IL-10, IL-1RA, IFN-γ, IL-13, MCP-1, IL-7, IL-15, IFN-α, IL-2R, IP-10, IL-5, and IL-2.
The data were acquired on the Luminex 100 System and were analyzed by a standard curve fit, according to the manufacturer’s protocol (72). In brief, for each analyte, a 7-point dilution series of the protein standard plus an assay diluent-only background well were run in duplicate. A standard curve was then fitted using a weighted 5-parameter logistic function. Protein concentrations of the unknown samples were quantified from the standard curves, and duplicates were averaged for downstream statistical analysis. The standard curve modeling was performed in R version 3.2.0 (73).
Flow cytometry. Mock- or DENV-infected DCs were first incubated in Human TruStain FcX (Biolegend), followed by incubation using the Live/Dead Fixable Blue Dead Cell Stain Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Cells were then fixed and permeabilized with Cytofix/Cytoperm (BD Pharmingen) and subsequently stained with anti-DENV NS3 monoclonal antibody E1D8 (74) (a gift from Eva Harris, University of California, Berkeley, Berkeley, California, USA) at 10 μg/ml, which was directly conjugated with DyLight 488 (Thermo Fisher Scientific). Samples were run on an LSRII flow cytometer, available through Mount Sinai’s Flow Cytometry Shared Resource Facility. Data analysis was performed with Cytobank software version 5.2.0 (Cytobank Inc.).
CyTOF staining and analysis. All CyTOF reagents were acquired from Fluidigm Inc. unless otherwise indicated. All antibodies were either purchased preconjugated from Fluidigm or conjugated and validated in-house using MaxPar X8 conjugation kits (Fluidigm Inc.) according to the manufacturer’s instructions. Antibody clone numbers are provided in Supplemental Table 1. Viability staining was first performed on the mock- or DENV-infected DCs by adding 1 μM Rh103 nucleic acid intercalator directly to the cell culture media at the indicated time point after infection and incubating for 20 minutes at 37°C. Samples were then washed in PBS containing 0.1% BSA, blocked with Human TruStain FcX (Biolegend), and incubated with a 0.1-micron-filtered cocktail of titrated MaxPar antibodies against cell surface markers (Supplemental Table 1) for 20 minutes at 4°C. The samples were washed, and mock and DENV-infected samples were subsequently fixed and barcoded using a Cell-ID 20-Plex Pd barcoding kit with a unique barcode ID assigned to each condition and time point. After barcoding, the samples were pooled to minimize staining variability, permeabilized using BD Cytofix/Cytoperm (BD Biosciences), and incubated with a 0.1-micron-filtered cocktail of MaxPar antibodies against intracellular antigens (Supplemental Table 1) for 20 minutes at 4°C. The samples were then washed, incubated with 0.125 nM Ir nucleic acid intercalator to enable cell identification based on DNA content, and stored in PBS containing 2% freshly diluted formaldehyde (Electron Microscopy Sciences) until acquisition. Samples collected at the 8, 24, 48, and 72 hpi time points were stored and then combined together and acquired simultaneously as a single barcoded sample to minimize acquisition batch effects. Immediately prior to acquisition, the barcoded samples were washed in diH20 and resuspended at a concentration of 600,000 cells per ml with a 1:20 dilution of EQ 4 element beads. Samples were acquired on a CyTOF2 using a SuperSampler fluidics system (Victorian Airships) at an event rate of <350 events/s. Following data acquisition, data were concatenated and normalized using the CyTOF2 software. The barcoded samples were then deconvoluted using a Matlab-based debarcoding and doublet-filtering application (75), using a stringent Mahalanobis distance cutoff to ensure optimal barcode separation. The data were then uploaded to Cytobank version 5.2.0 (Cytobank Inc.) for analysis.
The data were analyzed using a combination of manual gating approaches (Supplemental Figure 3A), autogating based on 99th percentile baseline thresholding (Figure 2 and Supplemental Figure 3B) (36, 37), and viSNE (Figure 3 and Supplemental Figure 7) (35). Manual and automatic gating have comparative advantages and disadvantages that could be accentuated by the high dimensionality of CyTOF data. Although it is currently the standard method, manual gating may be subject to operator bias in choosing thresholds for marker positivity or negativity (76), whereas autogating is not subject to such bias, as it specifies a deterministic and perfectly reproducible threshold for each channel. On the other hand, the sensitivity and specificity of autogating may be affected by data quality and certain analytical assumptions, such as the unexpected presence of outlier data that could skew automatically calculated thresholds (77), whereas an expert human operator would be able to notice and correct for this problem. By utilizing both types of analyses in this study, the patterns of cell phenotypes were shown to be generally consistent under either method (Supplemental Figure 3B) and therefore robust to the choice of gating technique.
Additionally, viSNE analysis allowed for the visualization of high-dimensional, single-cell CyTOF data in two-dimensional plots (35). With this dimensionality-reducing algorithm, phenotypic changes could be appreciated in a coordinated pattern, in which the relative position of a cell on the plot indicated its general phenotype in multidimensional space (35). The staining intensities for the indicated markers were displayed as expression level gradients. viSNE plots were generated using all 18 phenotypic markers from the CyTOF panel (Supplemental Table 1). DENV E and NS3 proteins were purposefully excluded as clustering parameters in order to visualize the distribution of infection across cellular phenotypes.
Statistics. For data with multiple DC donors, significance was determined using a paired, 2-tailed Wilcoxon signed-rank test, which does not assume a normally distributed population. The Benjamini-Hochberg procedure was performed at each time point for each cytokine/chemokine or CyTOF marker to adjust the significance level for multiple comparisons. P ≤ 0.05 after correcting for multiple comparisons was considered statistically significant. Wilcoxon signed-rank tests were performed with Prism 7 GraphPad software, and Benjamini-Hochberg procedures were performed with R version 3.3.0.
Event counts for manually gated CyTOF data were exported from Cytobank Inc. Autogating of CyTOF data was performed by setting a threshold at the 99th percentile of the distribution of observed intensities for each marker at the 8 hpi time point in mock-treated samples (36, 37). This threshold was then applied to all other time points and treatments for that donor, which created event counts for positive and negative subpopulations. The comparative effects of infection condition on marker expression were calculated within each donor as the log10 of the relative risk for positive marker frequency in the live populations extracted at each time point for each marker. Since manual gating and autogating strategies were determined to show grossly consistent patterns (Supplemental Figure 3B), autogating was used for more extensive analyses (Figure 2). In these analyses, markers were ordered vertically by a hierarchical clustering of the median log10 relative risks for that marker and time point. Hierarchical clustering was performed using the R hclust function using “complete” agglomeration and the Euclidean distance between row vectors (73). Statistical significance was determined using a 2-sided Fisher’s exact test at a threshold of α = 10–5 with Bonferroni correction for multiple testing, similar to previous analyses of flow cytometry counts (78). Statistical analyses for Figure 2 were performed with R version 3.2.2 (73). Autogating was implemented using the flowCore (79) and flowStats (80) R packages from Bioconductor (versions 1.36.9 and 3.28.1, respectively).
Study approval. Primary human cells used in this work were obtained from buffy coats from the New York Blood Center and were deidentified prior to distribution by the New York Blood Center. As such, these samples are considered to be nonhuman subjects and do not require institutional review board approval.
-
Author contributions
REH, AR, TRP, KM, AK, IR, MM, and AFS designed the research studies. REH, AR, KM, IM, DBR, UP, AMM, and ACF conducted experiments and acquired the data. REH, AR, TRP, KM, IM, EDA, AK, IR, MM, and AFS analyzed the data. REH and AFS wrote the manuscript. REH, AR, TRP, KM, IM, AMM, ACF, AK, IR, MM, and AFS edited the final manuscript.
-
Acknowledgments
We thank Andrea Gamarnik, Eva Harris, and Raul Andino for providing reagents and for insightful discussions. We also thank the Human Immune Monitoring Core at Mount Sinai for technical assistance and John Bucci at Thermo Fisher Scientific for assistance with multiplex ELISA analysis. This study has been partially funded by NIH/National Institute of Allergy and Infectious Diseases grants 1U19AI118610 (to AFS, MM, and AK), R01AI073450 (to AFS), 1R21AI116022 (to AFS), F30AI114161 (to REH), 5T32AI007647 (to REH), F30AI122673 (to TRP), and T32GM007280 (to REH and TRP); Defense Advanced Research Projects Agency (Prophecy) grant HR0011-11-C-0094 (to AFS); and a Sir Henry Wellcome fellowship 096062 (to KM).
Address correspondence to: Ana Fernandez-Sesma, Icahn School of Medicine at Mount Sinai, Department of Microbiology, Box 1124, One Gustave L. Levy Place, New York, New York 10029, USA. Phone: 212.241.5182; Email: ana.sesma@mssm.edu.
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information: JCI Insight. 2017;2(13):e92424. https://doi.org/10.1172/jci.insight.92424.
-
References
- Deen JL, et al. The WHO dengue classification and case definitions: time for a reassessment. Lancet. 2006;368(9530):170–173.
- Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507.
- Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. 2nd edition. World Health Organization. http://www.who.int/csr/resources/publications/dengue/Denguepublication/en/ Accessed June 19, 2017.
- Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition. World Health Organization. http://www.who.int/rpc/guidelines/9789241547871/en/ Accessed June 19, 2017.
- Dengue and severe dengue: fact sheet. World Health Organization. http://www.who.int/mediacentre/factsheets/fs117/en/ Accessed May 23, 2017.
- Fu J, Tan BH, Yap EH, Chan YC, Tan YH. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90). Virology. 1992;188(2):953–958.
- Vaughn DW, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181(1):2–9.
- Nishiura H, Halstead SB. Natural history of dengue virus (DENV)-1 and DENV-4 infections: reanalysis of classic studies. J Infect Dis. 2007;195(7):1007–1013.
- Fried JR, et al. Serotype-specific differences in the risk of dengue hemorrhagic fever: an analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoS Negl Trop Dis. 2010;4(3):e617.
- Thomas L, et al. Clinical presentation of dengue by serotype and year of epidemic in Martinique. Am J Trop Med Hyg. 2014;91(1):138–145.
- Thomas L, et al. Influence of the dengue serotype, previous dengue infection, and plasma viral load on clinical presentation and outcome during a dengue-2 and dengue-4 co-epidemic. Am J Trop Med Hyg. 2008;78(6):990–998.View this article via: PubMed Google Scholar
- De Carvalho Bittencourt M, Martial J, Cabié A, Thomas L, Césaire R. Decreased peripheral dendritic cell numbers in dengue virus infection. J Clin Immunol. 2012;32(1):161–172.
- Pham AM, Langlois RA, TenOever BR. Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 2012;8(1):e1002465.
- Durbin AP, et al. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376(2):429–435.
- Kyle JL, Beatty PR, Harris E. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J Infect Dis. 2007;195(12):1808–1817.
- Rodriguez-Madoz JR, Bernal-Rubio D, Kaminski D, Boyd K, Fernandez-Sesma A. Dengue virus inhibits the production of type I interferon in primary human dendritic cells. J Virol. 2010;84(9):4845–4850.
- Aguirre S, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8(10):e1002934.
- Ho LJ, et al. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J Immunol. 2001;166(3):1499–1506.
- Libraty DH, Pichyangkul S, Ajariyakhajorn C, Endy TP, Ennis FA. Human dendritic cells are activated by dengue virus infection: enhancement by gamma interferon and implications for disease pathogenesis. J Virol. 2001;75(8):3501–3508.
- Marovich M, et al. Human dendritic cells as targets of dengue virus infection. J Investig Dermatol Symp Proc. 2001;6(3):219–224.
- Wu SJ, et al. Human skin Langerhans cells are targets of dengue virus infection. Nat Med. 2000;6(7):816–820.
- Fujii S, Liu K, Smith C, Bonito AJ, Steinman RM. The linkage of innate to adaptive immunity via maturing dendritic cells in vivo requires CD40 ligation in addition to antigen presentation and CD80/86 costimulation. J Exp Med. 2004;199(12):1607–1618.
- Lee AW, et al. A clinical grade cocktail of cytokines and PGE2 results in uniform maturation of human monocyte-derived dendritic cells: implications for immunotherapy. Vaccine. 2002;20 Suppl 4:A8–A22.View this article via: PubMed Google Scholar
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392(6673):245–252.
- Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, Fernandez-Sesma A. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J Virol. 2010;84(19):9760–9774.
- Srikiatkhachorn A, Green S. Markers of dengue disease severity. Curr Top Microbiol Immunol. 2010;338:67–82.View this article via: PubMed Google Scholar
- Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. 2011;11(8):532–543.
- Gunther VJ, et al. A human challenge model for dengue infection reveals a possible protective role for sustained interferon gamma levels during the acute phase of illness. Vaccine. 2011;29(22):3895–3904.
- Sierra B, et al. Variation in inflammatory/regulatory cytokines in secondary, tertiary, and quaternary challenges with dengue virus. Am J Trop Med Hyg. 2012;87(3):538–547.
- Malavige GN, Huang LC, Salimi M, Gomes L, Jayaratne SD, Ogg GS. Cellular and cytokine correlates of severe dengue infection. PLoS One. 2012;7(11):e50387.
- Ashour J, Laurent-Rolle M, Shi PY, García-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83(11):5408–5418.
- Muñoz-Jordan JL, Sánchez-Burgos GG, Laurent-Rolle M, García-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100(24):14333–14338.
- Chase AJ, Medina FA, Muñoz-Jordán JL. Impairment of CD4+ T cell polarization by dengue virus-infected dendritic cells. J Infect Dis. 2011;203(12):1763–1774.
- Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696.
- Amir el-AD , et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552.
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152.
- Finak G, et al. OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput Biol. 2014;10(8):e1003806.
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516.
- Muñoz-Jordán JL, et al. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79(13):8004–8013.
- Ho LJ, et al. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J Immunol. 2005;174(12):8163–8172.
- Hannemann H, et al. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J Biol Chem. 2013;288(31):22621–22635.
- Kumar A, et al. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J Virol. 2013;87(8):4545–4557.
- Tay MY, et al. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013;99(3):301–306.View this article via: PubMed Google Scholar
- Welsch S, et al. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–375.
- De Maio FA, et al. The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog. 2016;12(8):e1005841.
- Villordo SM, Carballeda JM, Filomatori CV, Gamarnik AV. RNA Structure Duplications and Flavivirus Host Adaptation. Trends Microbiol. 2016;24(4):270–283.
- Manokaran G, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350(6257):217–221.
- Göertz GP, Pijlman GP. Dengue Non-coding RNA: TRIMmed for Transmission. Cell Host Microbe. 2015;18(2):133–134.
- Filomatori CV, et al. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog. 2017;13(3):e1006265.
- Palmer DR, et al. Differential effects of dengue virus on infected and bystander dendritic cells. J Virol. 2005;79(4):2432–2439.
- Nightingale ZD, Patkar C, Rothman AL. Viral replication and paracrine effects result in distinct, functional responses of dendritic cells following infection with dengue 2 virus. J Leukoc Biol. 2008;84(4):1028–1038.
- Dejnirattisai W, et al. A complex interplay among virus, dendritic cells, T cells, and cytokines in dengue virus infections. J Immunol. 2008;181(9):5865–5874.
- Sun P, et al. CD40 ligand enhances dengue viral infection of dendritic cells: a possible mechanism for T cell-mediated immunopathology. J Immunol. 2006;177(9):6497–6503.
- Wu WL, Ho LJ, Chang DM, Chen CH, Lai JH. Triggering of DC migration by dengue virus stimulation of COX-2-dependent signaling cascades in vitro highlights the significance of these cascades beyond inflammation. Eur J Immunol. 2009;39(12):3413–3422.
- Olagnier D, et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog. 2014;10(12):e1004566.
- Sanchez V, Hessler C, DeMonfort A, Lang J, Guy B. Comparison by flow cytometry of immune changes induced in human monocyte-derived dendritic cells upon infection with dengue 2 live-attenuated vaccine or 16681 parental strain. FEMS Immunol Med Microbiol. 2006;46(1):113–123.
- Taweechaisupapong S, et al. Langerhans cell density and serological changes following intradermal immunisation of mice with dengue 2 virus. J Med Microbiol. 1996;45(2):138–145.
- Taweechaisupapong S, Sriurairatana S, Angsubhakorn S, Yoksan S, Bhamarapravati N. In vivo and in vitro studies on the morphological change in the monkey epidermal Langerhans cells following exposure to dengue 2 (16681) virus. Southeast Asian J Trop Med Public Health. 1996;27(4):664–672.View this article via: PubMed Google Scholar
- Schmid MA, Harris E. Monocyte recruitment to the dermis and differentiation to dendritic cells increases the targets for dengue virus replication. PLoS Pathog. 2014;10(12):e1004541.
- Zhu X, et al. IFITM3-containing exosome as a novel mediator for anti-viral response in dengue virus infection. Cell Microbiol. 2015;17(1):105–118.
- Chahar HS, Bao X, Casola A. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses. 2015;7(6):3204–3225.
- Aline F, Bout D, Amigorena S, Roingeard P, Dimier-Poisson I. Toxoplasma gondii antigen-pulsed-dendritic cell-derived exosomes induce a protective immune response against T. gondii infection. Infect Immun. 2004;72(7):4127–4137.
- Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110(9):3234–3244.
- Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J Biol Chem. 2007;282(35):25779–25789.
- Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J Virol. 2000;74(17):7814–7823.
- Halstead SB, Simasthi P. Observations related to the pathogenesis of dengue hemorrhagic fever. II. Antigenic and biologic properties of dengue viruses and their association with disease response in the host. Yale J Biol Med. 1970;42(5):276–292.View this article via: PubMed Google Scholar
- Lanciotti RS, Gubler DJ, Trent DW. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78(Pt 9):2279–2284.View this article via: PubMed Google Scholar
- Halstead SB, Marchette NJ. Biologic properties of dengue viruses following serial passage in primary dog kidney cells: studies at the University of Hawaii. Am J Trop Med Hyg. 2003;69(6 Suppl):5–11.View this article via: PubMed Google Scholar
- Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74(11):4957–4966.
- Pickett BE, et al. Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses. 2012;4(11):3209–3226.View this article via: PubMed Google Scholar
- Haye K, Burmakina S, Moran T, García-Sastre A, Fernandez-Sesma A. The NS1 protein of a human influenza virus inhibits type I interferon production and the induction of antiviral responses in primary human dendritic and respiratory epithelial cells. J Virol. 2009;83(13):6849–6862.
- Gottschalk PG, Dunn JR. The five-parameter logistic: a characterization and comparison with the four-parameter logistic. Anal Biochem. 2005;343(1):54–65.
- R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ Accessed May 23, 2017.
- Balsitis SJ, et al. Tropism of dengue virus in mice and humans defined by viral nonstructural protein 3-specific immunostaining. Am J Trop Med Hyg. 2009;80(3):416–424.View this article via: PubMed Google Scholar
- Zunder ER, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10(2):316–333.
- Lugli E, Roederer M, Cossarizza A. Data analysis in flow cytometry: the future just started. Cytometry A. 2010;77(7):705–713.View this article via: PubMed Google Scholar
- Aghaeepour N, et al. Critical assessment of automated flow cytometry data analysis techniques. Nat Methods. 2013;10(3):228–238.
- Horton H, et al. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323(1):39–54.
- Hahne F, et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. View this article via: PubMed Google Scholar
- Hahne F GN, Khodabakhshi AH, Wong C, Lee K. flowStats: Statistical methods for the analysis of flow cytometry data. R Packages. https://rdrr.io/bioc/flowStats/ Accessed May 23, 2017.
-
Version history
- Version 1 (July 6, 2017): Electronic publication
- Version 2 (September 15, 2017): ImmPort accession numbers added.









