Research ArticleImmunologyVirology
Open Access |  10.1172/jci.insight.188543
10.1172/jci.insight.188543
Early multiple sclerosis activity associated with TBX21+CD21loCXCR3+ B cell expansion resembling EBV-induced phenotypes
Elliott D. SoRelle,1 Ellora Haukenfrers,2 Gillian Q. Horn,3 Vaibhav Jain,2 James Giarraputo,4 Karen Abramson,2 Emily Hocke,2 Laura A. Cooney,5,6 Kristina M. Harris,5 Scott S. Zamvil,5,7 Simon G. Gregory,1,2,8 and Micah A. Luftig1
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by SoRelle, E. in: PubMed | Google Scholar
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by Haukenfrers, E. in: PubMed | Google Scholar
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by Horn, G. in: PubMed | Google Scholar
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by
Jain, V.
in:
PubMed
|
Google Scholar
|

1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by Giarraputo, J. in: PubMed | Google Scholar
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by Abramson, K. in: PubMed | Google Scholar
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by Hocke, E. in: PubMed | Google Scholar
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by Cooney, L. in: PubMed | Google Scholar
1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by
Harris, K.
in:
PubMed
|
Google Scholar
|

1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by
Zamvil, S.
in:
PubMed
|
Google Scholar
|

1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by
Gregory, S.
in:
PubMed
|
Google Scholar
|

1Department of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, North Carolina, USA.
2Duke Molecular Physiology Institute, Duke University, Durham, North Carolina, USA.
3Department of Integrative Immunobiology, Duke University School of Medicine, Durham, North Carolina, USA.
4Yale School of Medicine, New Haven, Connecticut, USA.
5Immune Tolerance Network, Bethesda, Maryland, USA.
6Department of Internal Medicine, Division of Rheumatology, University of Michigan, Ann Arbor, Michigan, USA.
7Department of Neurology, University of California, San Francisco, San Francisco, California, USA.
8Department of Neurology, Duke University School of Medicine, Durham, North Carolina, USA.
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
Authorship note: SGG and MAL contributed equally to this work.
Find articles by
Luftig, M.
in:
PubMed
|
Google Scholar
|

Authorship note: SGG and MAL contributed equally to this work.
Published May 13, 2025 - More info
JCI Insight. 2025;10(12):e188543. https://doi.org/10.1172/jci.insight.188543.
© 2025 SoRelle et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: October 29, 2024; Accepted: April 30, 2025
-
Abstract
Epstein-Barr virus (EBV) infection precedes multiple sclerosis (MS) onset and plays a poorly understood etiologic role. To investigate possible viral pathogenesis, we analyzed single-cell expression in peripheral B cells from people with early MS collected longitudinally during the Immune Tolerance Network STAyCIS Trial. Expression profiles were compared with single-cell RNA-Seq (scRNA-Seq) from in vitro EBV models, autoimmune disorders, chronic infectious diseases, and healthy controls. Analyses focused on CD19+CD20+CD21loCD11c+T-bet+ atypical B cells (ABCs). ABCs were significantly enriched in early MS PBMCs versus healthy controls by scRNA-Seq and flow cytometry, establishing ABC expansion as a clinical feature. EBV-associated ABC expression, including CXCR3, programmed cell death ligand 1 (PD-L1), and PD-L2, was enriched in early MS; however, direct EBV infection of ABCs was not detected. Early MS ABCs exhibited significantly upregulated inflammatory cytokine mRNAs (CXCL8, IL18, VEGFA). Further, de novo EBV-infected B cells secreted IL-8 and VEGF. MS activity stratification revealed rare, distinctive inflammatory ABCs significantly underrepresented in individuals with no evidence of activity long-term versus people with additional relapsing-remitting MS activity at the primary endpoint. Moreover, CXCR3+ ABCs increased after baseline diagnosis and were significantly enriched in people with disease exacerbation during the study. Thus, ABC expansion and inflammatory responses correlate to early MS activity, possibly as a bystander response to EBV.
-
Introduction
The atypical B cell (ABC) compartment comprises immune cells coexpressing the T cell lineage transcription factor T-bet (TBX21), the plasmacytoid dendritic cell marker CD11c (ITGAX), and the classical B cell markers CD19 and CD20 (MS4A1) but lacking CD21, CD27, and IgD (1, 2). Originally discovered in mice as age associated, these cells accumulate in response to Toll-like receptor 7 (TLR7) and interferon-γ (IFN-γ) stimulation and exhibit B cell receptor anergy in addition to molecular indicators of exhaustion and germinal center–independent (GC-independent) development (1, 3–7). ABC pools contain high frequencies of autoreactive clones and are expanded in females due at least partially to dosage effects of TLR7, which is encoded on the X chromosome and escapes X-linked inactivation (1, 8–11). Although present in healthy individuals and capable of mediating protective immune responses (12–14), T-bet+ B cells coexpressing combinations of other markers (e.g., Fc receptor like 5 [FCRL5]/CXCR3) are commonly pathogenic activated effectors in autoimmune and infectious diseases with female bias (15, 16). These include systemic lupus erythematosus (SLE) (9, 17–19), primary Sjögren’s syndrome (PSS) (20), rheumatoid arthritis (21, 22), multiple sclerosis (MS) (23), and chronic infections with HIV and malaria (MAL) (24–29).
Epstein-Barr virus (EBV) is a gammaherpesvirus establishing lifelong infection in more than 90% of adults worldwide (30). This prevalence is attributable to the success with which EBV induces and modulates adaptive immune programs of host B cells to achieve persistent latency in the memory B cell pool via GC-dependent and GC-independent routes (31–34). Beyond its carcinogenic potential (35), EBV is frequently associated with autoimmune and chronic infectious diseases that exhibit clonally expanded ABCs (36–47). We recently reported that EBV can infect existing ABCs (34) and promote ABC generation in vitro (48). However, the manifestation and mechanistic contributions of EBV infection in autoimmune and chronic infectious diseases remain incompletely understood.
MS is a chronic neuroinflammatory disease with a 3:1 female bias (23) in which both pathogenic ABCs (49, 50) and epidemiologic evidence of EBV etiology in disease onset (a 32-fold risk from EBV seropositivity after childhood) have been identified (51, 52). Several hypotheses have been proposed to explain EBV involvement in MS: immune responses to viral proteins cross-reactive with self-antigens (molecular mimicry), EBV-induced licensing of forbidden autoreactive B cell clones, and inflammatory damage from infected cells (53–55). Antibody reactivity and T cell profiles from people with MS show molecular mimicry between the viral latency protein EBV nuclear antigen 1 (EBNA1) and CNS-expressed host proteins (56, 57). Multiple EBV antigens and EBV+ B cells have been detected in MS brain lesions though inconsistently (58–63). Notably, a correlation between neuroinvasive CXCR3+ B cells and EBV viral load exists in patients with MS (64). Beyond shedding light on the clinical efficacy of B cell depletion therapy, these findings underscore immune dysregulation mediated by EBV in B cells within the CNS. How the virus accesses the CNS and promotes pathogenesis remains unclear, but clinical aspects of MS align with markers and functions of ABCs, suggesting EBV infection perturbs this immune niche.
This study applies single-cell sequencing to investigate B cells from multiple diseases, focusing on samples from the STAyCIS Trial (65), which studied atorvastatin efficacy in preventing relapses or new CNS lesions in individuals diagnosed with clinically isolated syndrome (CIS) per Controlled High-Risk Subjects Avonex Multiple Sclerosis Prevention Study criteria that led to the approval of intramuscular IFN-β1a (66). Patients diagnosed with MS by 2001 McDonald criteria were excluded (67), though CIS at the time would likely be diagnosed as early MS by current criteria; we use the terminology early MS (eMS) herein. CIS is an initial clinical episode of neurologic symptoms caused by neuroinflammation and demyelination often preceding MS (68). People with CIS presenting with brain or spinal cord lesions detected by MRI have 60%–80% chance of subsequent neurologic events and relapsing-remitting MS (RRMS) diagnosis (69–71). However, ABC characteristics in CIS (here, eMS) cohorts remain understudied, as does whether signatures of ABC response to EBV infection exist in eMS or across virus-associated diseases (34). To address these gaps, we analyzed ABCs from people with eMS and compared them with ABCs from other disease cohorts and in vitro EBV infection.
-
Results
Study population and procedures. Peripheral blood mononuclear cells (PBMCs) were collected from a subset of the original Immune Tolerance Network (ITN) STAyCIS Trial cohort of people (n = 16 of 81 original participants; 11 female, 5 male) at 2 time points following initial diagnosis: t1 (baseline visit; at least 28 days after completion of corticosteroid treatment and within 210 days of presentation) and t2 (at least 3 months post-t1; mean = 172 days post-t1, range = 84–357 days). Beginning at t1, 11 people were treated with atorvastatin and 5 were given a placebo as part of the ITN STAyCIS Trial (65) (ClinicalTrials.gov NCT00094172). Treatments were given for 12 months or until participants met the primary endpoint of MS activity, defined as 3 or more new T2 lesions or 1 clinical exacerbation. Eight of the 16 participants for whom samples were analyzed met the primary endpoint within the 12-month treatment window, and 2 additional participants met the primary endpoint in a follow-up period between 12 and 18 months. Of the remaining 6 participants, 3 were subsequently diagnosed with MS based on new T2 lesions. Thus, 13 of 16 participants in the analyzed cohort were diagnosed with MS based on the accepted McDonald criteria at that time. The remaining 3 participants were characterized post hoc as long-term no evidence of activity (LTNA) based on the collective absence of new T2 lesions, Gd-enhancing lesions, and clinical exacerbations in the 18 months following t1. Of the 13 individuals with additional after-baseline MS activity, 10 received atorvastatin and 3 received placebo; of the 3 LTNA individuals, 2 received atorvastatin and 1 received placebo. In the original trial, atorvastatin significantly reduced development of new T2 lesion activity but did not meet the combined imaging and clinical primary endpoint (65). Notwithstanding, the longitudinal cohort provides a unique window into the peripheral lymphocyte compartment in eMS stages.
PBMCs from each individual and time point were prepared as single-cell libraries (single-cell RNA-Seq [scRNA-Seq] + cellular indexing of transcriptomes and epitopes by sequencing [CITE-Seq]), which were sequenced, quality-controlled, and aligned to generate count matrices. Single-cell data from the eMS cohort were then analyzed in conjunction with publicly available scRNA-Seq datasets from autoimmune disorders (MS, SLE, PSS) (49, 72–74) and chronic infectious diseases (HIV, MAL) (27) in which pathogenic ABCs have been described, as well as healthy controls (Figure 1A). Peripheral B cells (naive, intermediate, memory, and plasmablasts) were identified from each dataset by cell type prediction with Azimuth (75) and isolated for downstream analysis. We also incorporated the analysis of ABCs prior and subsequent to EBV infection in vitro (34) to assess transcriptomic similarities among ABCs from disease and in vitro EBV infection. The eMS dataset contained 14,969 annotated B cells collected from individuals across the 2 time points (n = 32). Total B cells and library sizes for all datasets are provided as supplemental material (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.188543DS1). B cells from the eMS cohort and publicly available datasets were integrated using single-cell batch correction methods (75) into a single object containing about 57,000 cells. Unbiased clustering was applied to refine high-resolution B subsets via biomarker analysis and literature-based annotation (Figure 1B). Analysis of integrated B cells by dataset verified successful correction of batch and sequence depth variation (Figure 1C and Supplemental Figures 1 and 2).
 Figure 1
Figure 1ABCs are enriched in people with eMS who experience subsequent MS activity. (A) Overview of sample collection, analysis workflow, and dataset integration from eMS cohort with publicly available data. (B) Integrated uniform manifold approximation and projection (UMAP) by B cell subsets. Rest NB, resting naive B cells (yellow); Rest MB, resting memory B cells (light purple); Homeo NB, homeostatic naive B cells (orange); Act SMB, activated switched memory B cells (powder blue); Trans B, transitional B cells (red); Late Act SMB, late activated switched memory B cells (beige); ABC 1, atypical B cell phenotype 1 (dark gray); MZ/NSM B, marginal zone/non-switched memory B cells (green); ISG B, interferon-stimulated gene signature B cells (pink); CD5+ B, CD5+ B cells (blue); PB, plasmablasts (peach); AHEM B, B cells with upregulated autophagy, ATP hydrolysis, and epigenetic modifier expression (dark purple); ITGB3+ B, ITGB3+ITGA2B+ B cells (goldenrod); ABC 2, atypical B cell phenotype 2 (fuchsia). (C) Integrated UMAP by dataset. aHD, adult healthy donor (eggshell white); cHD, child healthy donor (light blue); eMS, early MS (medium blue); MS, multiple sclerosis (cream); aSLE, adult systemic lupus erythematosus (gray); cSLE, child systemic lupus erythematosus (dark blue); HIV, chronic HIV infection (gold); MAL, chronic malaria (bronze); PSS, primary Sjögren’s Syndrome (black). (D) B cell subset composition across conditions. (E) B cell subset composition by condition, normalized by dataset sample size. (F) Statistical comparisons of B subset frequencies in aHDs, all eMS, and eMS stratified by LTNA (seafoam green) and SMSA (magenta) outcomes. Two-sided Wilcoxon rank-sum test (*P < 0.05; **P < 0.01; ***P < 0.001). (G) Scaled expression of B subset marker genes. (H) TBX21 and CXCR3 expression.
Characterization of B cell populations within study cohort compared with chronic infection, autoimmunity, and healthy controls. Percentages of B cells within annotated subsets by disease were assessed (Figure 1D), verifying findings in prior studies such as ABC expansion (ABC 1) in SLE, HIV, and MAL; B cells with ISG signature (ISG B) in SLE; and PB enrichment in HIV and MAL. Generally, B cell compartments in chronic infections and autoimmune diseases were skewed toward lower frequencies of resting naive and memory B cells and higher frequencies of activated effector phenotypes (e.g., activated switched memory B cells, PBs). The proportion of cells from each disease context and healthy controls for each annotated B subset was quantified after normalizing for total cell numbers per dataset (Figure 1E). This representation revealed enrichment of comparatively rare B cell phenotypes in disease versus healthy controls. Beyond PB predominance from HIV, MAL, and eMS, we identified overrepresentation of eMS- and PSS-derived B cells with gene ontology (GO) enrichment for AHEM B; MS- and PSS-derived ITGB3+ B; and eMS- and PSS-derived ABC 2, an apparent, distinct, rare population of ABCs (based on coexpression of TBX21, CD19, MS4A1, and ITGAX). Statistical comparisons identified significant enrichment of ABC 1 (2-sided Wilcoxon rank-sum test, P = 0.031) and PB (Wilcoxon P = 1.55 × 10–4) in eMS versus healthy adults. Conversely, B cells distinguished by elevated expression of CD5 (CD5+ B) were less frequent in patients with eMS versus healthy controls (Wilcoxon P = 2.71 × 10–3). Stratification of the eMS cohort by subsequent MS activity (eMS→SMSA) versus individuals with no additional evidence of MS activity (eMS→LTNA) revealed differences in frequencies of late activated switched memory B cells (Late Act SMB; aHD < eMS→SMSA, Wilcoxon P = 0.042; eMS→LTNA < eMS→SMSA, Wilcoxon P = 2.72 × 10–3) and AHEM B cells (aHD < eMS→SMSA, Wilcoxon P = 0.036). Samples from eMS→SMSA displayed elevated ABC 2 frequencies relative to eMS→LTNA samples, though not statistically significant (Wilcoxon P = 0.07) (Figure 1F and Supplemental Figure 3A). Notably, no significant differences in B subset frequencies existed between atorvastatin versus placebo groups (Supplemental Figure 3B).
Next, we assessed differential gene expression for each annotated subset across integrated B cell datasets (Figure 1G and Supplemental Tables 2 and 3). Conventional naive and memory B subsets (Rest NB, Rest MB, Homeo NB) were identified by patterns of CD19, FCER2 (CD23), CD24, CD27, CD38, IGHD, IGHG1, and other B cell lineage markers. Trans B cells were distinguished by elevated CD9 and MME (CD10) expression. Activated B subsets were further delineated by expression of CD86, FAS (CD95), and CD80. In addition to elevated expression of TBX21 (T-bet), ITGAX (CD11c), and FCRL5, ABC 1 cells were distinguished by FOXP4, SOX5, TNFRSF1B, near-exclusive expression of AIRE, and low expression of CR2 (CD21). ISG B cells displayed the highest levels of IFI6, IFI16, IFI27, and ISG15. PBs expressed high SPN (CD43), SLAMF7, PRDM1 (BLIMP-1), and SDC1 (CD138). The CD5+ B phenotype also expressed CD6, CD2, MYB, and TNFRSF25, while ITGB3+ B cells expressed ITGA2B and moderate levels of CD9. AHEM B cells displayed elevated mitochondrial gene expression, sex-specific expression of XIST and its regulator FTX, and the epigenetic regulators HDAC6 and KMT2D. Finally, the ABC 2 cluster expressed inflammatory cytokine transcripts (IL1B and IL18) in addition to atypical B lineage genes (Figure 1G and Supplemental Figure 4). An examination of TBX21 and CXCR3 by B subset and dataset revealed that these genes were generally expressed at higher levels and in more B subsets in disease contexts versus healthy adults. Specifically, CXCR3 expression in ABC 1 and Late Act SMB subsets was significantly higher in eMS and MS samples versus healthy controls (Figure 1H). ABC 2 expression of CXCR3 and TBX21 was also elevated in eMS versus healthy controls, though only CXCR3 enrichment was significant by statistical test (CXCR3 Wilcoxon P = 0.043; TBX21 Wilcoxon P = 0.11) (Figure 1H). Variation in CXCR3 expression across diseases may reflect differential enrichment of B cells originating from distinct cell lineages (76) or changes in the balance of resting (CXCR3–/lo) versus stimulated (CXCR3hi) phenotypes (48, 77, 78). Collectively, scRNA-Seq resolved peripheral B cell landscape variations in health and disease and defined the expansion of CXCR3+ B subsets, including TBX21+CD21lo ABCs, as clinical features in people with eMS.
EBV-induced gene expression signature is enriched in eMS patient ABCs. Based on the positive association between CNS-infiltrating CXCR3+ B cells and EBV viral load in people with MS (64) and our prior identification of EBV-induced CXCR3 expression (34, 48), we next evaluated expression of EBV-induced gene expression signatures within the integrated B cell scRNA-Seq data (Supplemental Table 4, Figure 2, and Supplemental Figures 5–9). Using scRNA-Seq from in vitro EBV infection, we generated the top 100 DEGs in distinct EBV-induced phenotypes, including a comparison of EBV+ versus EBV– ABCs (Figure 2A and Supplemental Figure 5, A–C). The EBV+ ABC module score was highest in the ABC 1 subset as expected, with elevated scores observed for ABCs from all autoimmune diseases and chronic infections relative to healthy control ABCs (Figure 2, B–D). The EBV+ NF-κB activation signature was high in multiple activated effector subsets and strongest in Late Act SMB cells, though this module did not exhibit clear patterns between healthy and disease contexts (Supplemental Figure 5A). The PB gene module and curated ISG signature were elevated, respectively, in PB and ISG B subsets as expected, with higher ISG scores observed in eMS, SLE, HIV, and PSS (but not MS or MAL) relative to healthy adults (Supplemental Figure 5, B and C, and Supplemental Figure 6). Although GO results indicated similar biological process enrichment in Late Act SMBs and ABCs in eMS (Supplemental Figure 7), we focused our analysis on the 2 ABC subsets. One of these (ABC 1) was more frequent in people with eMS regardless of disease outcome and time point, while the other (ABC 2) was elevated in people with subsequent MS activity (eMS→SMSA). ABC 1 and ABC 2 subsets from people with eMS exhibited higher scores for genes in the top 100 EBV+ ABC DEGs when compared with ABCs from healthy donors (Supplemental Tables 5 and 6). These included CXCR3, B4GALT1, CLEC4A, TBKBP1, CREB5, NRCAM, CD274 programmed cell death ligand 1 (PD-L1), WASF3, PPP1R17, HIST1H3F, DLG4, and GPR174. Other EBV+ ABC markers (not within the top 100 DEGs) elevated in one or both eMS ABC subsets versus healthy donors included IL18, CMC4, and IFNG. Several of these EBV-associated genes expressed in ABCs were also upregulated in other disease contexts (Figure 2E and Supplemental Figure 8).
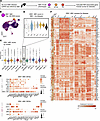 Figure 2
Figure 2EBV-associated gene expression signatures by B subset, disease, and clinical outcome. (A) Strategy to assess single-cell gene expression signatures associated with de novo EBV infection of B cells in vitro. ABC, EBV+ atypical B cell; NFκB, EBV+ NF-κB activated B cell phenotype; PB, EBV+ plasmablast phenotype. (B) UMAP representation of the in vitro EBV+ ABC gene signature across all integrated B cells from disease and healthy control datasets. DEGs, differentially expressed genes. (C) EBV+ ABC signature module scores by B cell state. Scores were calculated from the top 100 DEGs in EBV+ ABCs versus resting ABCs in vitro. (D) EBV+ ABC signature module scores in the ABC 1 cluster stratified by disease and healthy control datasets. (E) Dot plots of EBV+ ABC biomarker expression across ABC 1 (top panel) and ABC 2 (bottom panel) subsets in disease cohorts and healthy adults. (F) EBV+ ABC biomarker expression by dataset. Single-cell expression heatmap of ABC biomarkers and EBV-induced genes in ABCs during in vitro early infection. Genes (rows) are clustered by expression pattern similarity across datasets. Resting (EBV–) and infected (EBV+) ABCs from in vitro experiments are outlined in black and green boxes, respectively. EBV+ ABC gene groups identified by unbiased hierarchical ordering to have elevated expression in ABCs from patients with eMS but not healthy adults are outlined in cyan.
Hierarchical clustering of ABC lineage and EBV+ ABC genes refined single-cell expression patterns in ABC subsets by disease status (Figure 2F and Supplemental Figure 9). Genes including CD84, ENC1, FCGR2A (CD32), the EBV EBNA2 target FGR (79, 80), FOXP4, GAS7, GRAP2, HCK, MPP6, PLEK, and TNFRSF1B were consistently expressed by ABCs in most contexts, while certain genes were restricted to ABC clusters in one or more diseases but not healthy controls. Based on their functional importance to ABC responses, we examined genes with annotated roles in IFN-γ and TLR7 signaling. Indeed, FCRL3, PIK3AP1, SCIMP, and SLAMF6 were upregulated across ABCs. TLR7 expression, which is known to be induced upon EBV virion entry into B cells (81), was upregulated in ABCs from HIV, SLE, eMS, and MS versus healthy adults. Collectively, this analysis supported prior findings of ABC pathogenicity in autoimmunity and chronic infection, possibly mediated by common mechanisms (27, 82). The presence and extent of shared expression trends between disease state and EBV-infected ABCs further supported a potential association between EBV infection and pathogenic ABC expansion.
Expression of other EBV+ ABC genes including ALDH1A1, AREG, EDN1, SYTL4, TMEM63C, MCTP1, PPP1R17, and PRX was mostly specific to eMS and a subset of MS ABCs. Many of the EBV+ ABC genes not expressed in healthy controls play roles in development, cell plasticity, or neuronal lineage specification and were infrequently expressed in mature B subsets. It was striking that an idiosyncratic collection of genes upregulated in ABCs following EBV infection was also observed in disease-associated ABCs, but not healthy controls, and that these transcriptomic similarities were greatest in eMS. The lineage-ectopic nature of this signature may be due to unknown or uncharacterized functional roles for these gene products in B cells. Alternatively, it is conceivable that their expression, not normally detected in B cells, may be associated with induced reprogramming that promotes or maintains cellular plasticity (83–86). In this regard, we speculate that ABCs may be primed for aberrant responses to complex stimuli including viral infection.
EBV-regulated genes are commonly expressed in eMS/MS ABCs relative to healthy controls. Given these observations, we further examined the possibility of EBV-associated ABC pathogenicity (Figure 3 and Supplemental Figure 9B). ABC lineage markers, including CD19, MS4A1 (CD20), TBX21 (T-bet), and FCRL5, were conserved at similar levels in each ABC subset from healthy adults, eMS, MS, and in vitro EBV infection (Figure 3A). By contrast, significant upregulation of the immune checkpoint genes CD274 (PD-L1) and PDCD1LG2 (PD-L2) was observed in eMS, MS, and de novo EBV-infected ABCs versus healthy controls. The EBV+ ABC marker and MS susceptibility gene TNFRSF1A was significantly elevated in ABC 1 from people with eMS versus healthy adults (Wilcoxon P = 7.99 × 10–7). CD6 (another MS risk locus observed in EBV+ ABCs) was mildly elevated in ABC 2 from eMS, but not significantly (Wilcoxon P = 0.19), versus healthy controls (Figure 3B). Several genes upregulated in EBV+ and eMS ABCs are mediators of chemotaxis and tissue invasion, including the metalloproteinase ADAM9, which is induced in peripheral and tissue-infiltrating mononuclear cells under inflammatory conditions (87). While CXCR3 was expressed in ABC 1 from healthy donors, it was significantly upregulated in ABC 1 cells from people with eMS (Wilcoxon P = 4.58 × 10–9) and MS (Wilcoxon P = 4.6 × 10–14) and in ABC 2 cells from people with eMS (Wilcoxon P = 0.043). CXCR3 is notable because it facilitates migration from lymph nodes to inflammatory sites (88, 89) and has been defined as a characteristic of neuroinvasive B cells in diseases including MS (64, 90, 91). Because CXCR3+ B cell frequency in MS is positively correlated with EBV viral load (64) and the virus can induce B cell CXCR3 mRNA and protein expression (34, 48), infection may promote ABC trafficking to the CNS. WASF3, another enriched gene in ABC 1 from eMS and MS, encodes an actin-binding protein that regulates cytoskeletal morphology and promotes cellular metastasis and invasion in breast and prostate cancer (92, 93) (Figure 3C). Enrichment of numerous pro-inflammatory mediators (IL18, THBS3, VEGFA) associated with de novo EBV infection was also observed in ABC subsets from people with eMS (Figure 3D). In particular, ABC 2 cells were distinguished by elevated expression of genes encoding chemokine ligands, cytokines, and secreted factors with pro-inflammatory effects (Supplemental Figure 9B). These included CC motif chemokine ligands CCL3 (MIP-1A), CCL4 (MIP-1B), CCL5 (RANTES); CXC motif chemokine ligands CXCL2 (MIP-2A), CXCL3 (MIP-2B), CXCL8 (IL-8); granzymes GZMK, GZMM; IFNG (IFN-γ); IL18; IL1B; and TNF (TNF-α) — several of these have been previously identified in people with MS (94–96). CCL3, CCL4, CCL5, and IL18 were also significantly upregulated at the mRNA level during de novo EBV infection of ABCs. Elevated expression of IL18 and the pro-fibrotic wound-healing mediators AREG (97) and PDGFD (98) within EBV+ and eMS phenotypes further indicated potential functions or participation in (neuro)inflammatory responses. In this regard, expression of neuronal genes involved in cell-cell interactions and axon guidance (e.g., NRCAM, MCTP1, PPP1R17, SYTL4, TMEM63C) was further suggestive of neurotropism (Figure 3E). Upregulation of DLG4 in EBV+ and eMS ABCs was noteworthy, as this gene encodes a neuronal signaling mediator identified in MS leukocytes that was significantly downregulated during IFN-β treatment (99).
 Figure 3
Figure 3EBV-induced ABC markers observed in eMS ABCs are associated with pathogenic immune checkpoint evasion, neuroinvasive chemotaxis, and inflammatory responses. (A) ABC lineage marker expression within peripheral ABC 1 (gray) and ABC 2 (fuchsia) subsets across healthy adults (eggshell white), people with eMS (medium blue), people with MS (beige), and in vitro EBV infection (orange). (B) EBV+ ABC biomarkers: immune checkpoint and MS susceptibility gene expression color-coded by ABC phenotypes and dataset as in A. Statistically significant differences relative to healthy controls are represented by gray asterisks (ABC 1 phenotype) and fuchsia asterisks (ABC 2 phenotype) (Kolmogorov-Smirnov [KS] test, *P < 0.05; **P < 1 × 10–5; ***P < 1 × 10–10). (C) EBV+ ABC biomarkers: chemotactic and cell migration gene expression color-coded by ABC phenotypes and dataset as in A and B. Statistically significant differences in expression relative to healthy controls are represented as in B. (D) EBV+ ABC biomarkers: inflammatory mediator expression color-coded by ABC phenotypes and dataset as in A–C. Statistically significant differences in expression relative to healthy controls are represented as in B and C. (E) EBV+ ABC biomarkers: expression of lineage-ectopic neurotropic genes color-coded by ABC phenotypes and dataset as in A–D. Statistically significant differences in expression relative to healthy controls are represented as in B–D.
To investigate potential contributions of EBV infection to cytokine production and inflammatory responses, we performed a 27-plex Luminex assay comparing secretion profiles of resting peripheral B cells, early EBV-infected cells, and EBV-immortalized lymphoblastoid cell lines (LCLs). Secreted CCL2, CCL3, CCL4, CCL5, CXCL10, GM-CSF, IFN-γ, IL-8, IL-10, IL-1RA, TNF-α, and VEGF were all significantly elevated in de novo EBV-infected proliferative B cells relative to resting cells (2-sided Wilcoxon rank-sum tests, P < 0.05 for all). Secreted CCL3, CCL4, CCL5, CXCL10, IFN-γ, IL-10, and VEGF were also significantly elevated in LCLs but at lower levels than in de novo infection (Supplemental Figure 10).
We next applied GO enrichment to compare ABC 1 cells from outcome-stratified eMS samples versus healthy adults (Supplemental Figure 11, A and B). Regardless of outcome, top enriched terms for ABC 1 in the eMS cohort versus ABC 1 in healthy adults were related to viral transcription and mRNA catabolism, including nonsense-mediated decay, which mediates antiviral immunity in RNA and DNA virus infections (100–102). Despite these findings, we did not directly detect EBV viral transcripts in any B subset from people with eMS via read alignment against a multispecies reference genome. Thus, it cannot be definitively determined whether pathogenic features of ABC subsets in people with eMS detailed above are directly induced by EBV infection or represent ABC-specific responses that may be triggered indirectly by EBV infection or other stimuli.
ABC gene expression programs correlate with subsequent MS activity after baseline. We next extensively characterized differences in B cell composition, phenotypes, and ABC compartments between eMS→SMSA and eMS→LTNA groups. After accounting for different sample sizes, resting or homeostatic naive B cells (Rest NB, Homeo NB) were more frequent in eMS→LTNA samples. The ABC 1 subset was present at similar frequencies in SMSA and LTNA outcomes. The ISG B subset was more prevalent in the LTNA group; however, this was attributable to an individual sample. The ABC 2 phenotype was overrepresented in people with eMS who experienced subsequent MS disease activity (Supplemental Figure 12A). Cell-matched surface protein expression assayed via CITE-Seq verified that ABC 1 and ABC 2 subsets were CD19+CD11c+CD27lo (Supplemental Figure 12, B and C). Further, CD19 and CD11c expression were elevated in ABCs from eMS→SMSA samples. Intriguingly, ABC 2 cells (n = 37 cells, 36 of which were derived from eMS→SMSA outcomes) displayed broad immune lineage expression including CD11b (ITGAM), CD14, and CD64 (Supplemental Figure 12C). Likewise, ABC 2 cells exhibited an unusually broad set of surface-expressed genes in the scRNA-Seq assay, many of which were expressed at higher levels in eMS and MS versus healthy controls (Supplemental Figure 11C). The most distinctive of these included CD14, CD33, CD274 (PD-L1), HAVCR2 (CD366), ITGAM (CD11b), colony-stimulating factor receptor family members (CSF1R, CSF2RA, CSF3R), FcγR family members (FCGR1A [CD64], FCGR3A, FCGR3B), killer cell lectin-like receptors (KLRB1, KLRD1, KLRK1), and classic T cell markers (CD4, CD8A, CD8B) in addition to ABC hallmarks (CD19+, ITGAX+, and CR2lo). This broad mRNA immunophenotype included features — CD19+, PTPRC (CD45)+, FCER2 (CD23)lo, CR2 (CD21)lo, ITGA4 (VLA-4)+, ITGAL (LFA-1)+, TLR4 (CD284)+, CD5int — consistent with pro-inflammatory GM-CSF+ B cells described in MS (103), which are closely related but not identical to innate response activator B cells in mice (104). Given the breadth of the ABC 2 cluster expression profile, we considered whether these cells might be doublets. The numbers of unique features and read counts per cell in ABC 2 were comparable to other phenotypes, consistent with singlets (Supplemental Figure 1B). Consistent with Azimuth scoring, expression of CD19 and MS4A1 as well as B lineage transcription factors (e.g., PAX5, BACH2, EBF1) were characteristic of cells in ABC 2. Substantially fewer cells within the cluster expressed non-B lineage transcription factors (e.g., TCF7, GATA3, CEBPA, GATA2). Detailed examination of the raw RNA counts for highly expressed cell type biomarkers from eMS ABC 2 cells (n = 37 total) identified 28 cells with nonzero MS4A1 (CD20) versus 2 cells each with nonzero CD3E (1 cell also CD19+ and MS4A1+) and FCGR1A (1 cell also CD19+ and the other MS4A1+). Finally, reanalysis of the sequencing data with scDblFinder (v1.18; https://github.com/plger/scDblFinder; commit ID 20e4d51) identified only 1 of the 37 eMS cohort barcodes in ABC 2 as a potential doublet. Thus, the ABC 2 cluster had numerous characteristics to support assignment as singlet B cells, though limited presence of other cells may not be excluded definitively.
We next performed GO enrichment of differentially expressed markers between ABCs from eMS→SMSA and eMS→LTNA outcomes (Supplemental Tables 7 and 8). GO terms significantly enriched in eMS→SMSA ABCs were related to B cell activation, signal transduction, immune response, nucleotide biosynthesis, and cellular respiration. By contrast, GO terms enriched in eMS→LTNA ABCs were related to epigenetic modifications, mRNA splicing and transport, and regulation of cellular and mitochondrial autophagy (Supplemental Figure 13A). In addition to being elevated in eMS→LTNA versus eMS→SMSA, numerous genes within these GO categories were depleted in ABCs upon in vitro EBV infection (Supplemental Figure 13B). These included EZH2, BCOR, and JMJD6 (related to chromatin modification); DRB1, DDX39A, and WBP11 (related to RNA splicing); and SQSTM1, TMEM39A, and ULK1 (related to autophagy). Although not associated with any GO term, one of the top transcripts differentially upregulated in eMS→LTNA samples, MTRNR2L8, has previously been identified as a biomarker with potential predictive diagnostic value in MS, where reduced expression correlates with disease activity (105) (Supplemental Figure 7A).
Further examination of differential expression between SMSA and LTNA outcomes in ABC 1 and ABC 2 subsets highlighted differential pathogenic potential. ABCs from eMS→SMSA exhibited higher expression of genes involved in antiviral sensing (RIPK3, NOD1, TRIM34, TLR9, TLR4) and pro-inflammatory (OSM, IL1B, CXCL8) responses (Figure 4A and Supplemental Figure 14). Elevated NOD1, TLR9, and TLR4 expression in these cells supports an enhanced capacity for inflammatory responses to pathogen-associated molecular patterns. Similarly, RIPK3 and IL1B expression in eMS→SMSA ABCs was noteworthy given the role of RIPK3 in NLRP3 inflammasome activation and subsequent IL-1B activation (106, 107). By contrast, ABCs from eMS→LTNA samples were defined by elevated expression of genes with roles in limiting inflammatory responses (Figure 4B and Supplemental Figure 14). These included NR4A3, which antagonizes NF-κB signaling (108, 109). FOXO3 is essential for the induction of autophagy (110), which limits inflammatory responses by targeting activated inflammasomes (111). Moreover, NLRP6 expression was consistent with negative regulation of inflammation in the context of innate immune responses (112). GO analysis of genes differentially elevated in ABC 2 versus ABC 1 in people with eMS identified terms related to immune cell activation, cytotoxic degranulation, IL-1β and TNF cytokine production, and TLR signaling pathways (Supplemental Figure 15). Thus, expression in ABC subsets from eMS→SMSA versus eMS→LTNA, especially ABC 2 cells, support differential outcomes related to control of cellular inflammatory responses, possibly resulting from pathogenic stimulation. Despite the lack of detected EBV reads, stratifying ABCs from people with eMS according to clinical outcome (SMSA versus LTNA) revealed significantly higher expression of the EBV+ ABC DEG module in samples from individuals exhibiting subsequent disease activity. Moreover, EBV+ ABC module scores were significantly higher in t2 versus t1 sample ABCs from people with eMS who experienced SMSA but not in LTNA (Figure 4C). Numerous EBV-induced ABC DEGs including NUDT17, ALDH1A1, IFI27, and TNFRSF1A were enriched in eMS→SMSA versus eMS→LTNA (Figure 4D).
 Figure 4
Figure 4Outcome-stratified functional enrichment of ABCs from people with eMS implicates inflammatory responses linked to antiviral control in MS disease activity. (A) Dot plot of GO term–associated genes differentially upregulated in ABCs from eMS→SMSA versus eMS→LTNA. Expression is presented relative to expression in healthy adults (HD_t0). Select genes are denoted by arrows indicating their functional involvement. Dot size represents the percentage of ABCs expressing a gene. Dot color denotes scaled average gene expression. (B) Dot plot of GO term–associated genes upregulated in eMS→LTNA versus eMS→SMSA. Expression and gene function annotations are denoted as in A. (C) Time point– and outcome-stratified scoring of the top 100 EBV+ ABC DEGs in patients with eMS. Statistically significant differences in EBV+ ABC module score were evaluated by 2-sided Wilcoxon rank-sum test (*P < 0.05; ***P < 0.001). (D) Dot plot of select EBV+ ABC biomarkers (DEGs) stratified by time point and outcome in ABCs from people with eMS. Several ABC lineage genes are shown in addition to EBV-associated DEGs. Cell frequency and expression level are presented as in A and B.
While the ABC 2 cellular phenotype was rare, it was nearly 7-fold more frequent in eMS→SMSA than eMS→LTNA samples after accounting for sample sizes (~1 in 330 B cells in eMS→SMSA, ~1 in 2,500 B cells in eMS→LTNA). Overall, 36 cells from 8 of 13 people with eMS→SMSA activity exhibited the ABC 2 phenotype, compared with only 1 cell from 1 of 3 eMS→LTNA cases. Based on the number of ABC 2 cells expected from eMS→LTNA by scRNA-Seq library sizes, the probability of observing ≤1 cell by random chance alone was less than 1 in 50 (0.0082 < PPoisson < 0.0198; Supplemental Figure 16). This differential ABC 2 frequency between eMS→SMSA and eMS→LTNA further indicates ABC 2 contains singlets, since library prep and pooled sequencing of all eMS samples would minimize sample-dependent (outcome-dependent) doublet rates. Thus, the inflammation-associated ABC 2 phenotype was significantly underrepresented in LTNA versus people with subsequent MS activity.
We next used informatic methods to explore potential viral contribution to the observed eMS and EBV+ ABC phenotype. We used cis-regulatory prediction (113) to identify genes associated with EBNA binding sites detected using ChIP-Seq and cross-referenced these predictions against genes upregulated in eMS and EBV+ ABCs relative to resting cells (Supplemental Figure 17A). This yielded 82 EBNA-associated genes, including TBX21, CXCR3, FCRL2, FCRL5, FCGR2A, AREG, CCL5, CD69, CD84, CD300A, FGR, GPR174, HCK, and TBKBP1. These 82 genes constituted modest enrichment of GO terms including defense response (GO:0006952, n = 17 genes, FDR = 0.0442), leukocyte activation (GO:0045321, n = 14 genes, FDR = 0.0442), regulation of cell migration (GO:0030334, n = 13 genes, FDR = 0.0468), and leukocyte degranulation (GO:0043299, n = 10 genes, FDR = 0.0468). Analysis of the TBX21 locus in the LCL GM12878 revealed accessible chromatin with activating histone marks (H3K9ac, H3K4me3, H3K27ac), RNA polymerase II, and binding sites for EBNA-LP and EBNA2 at the TBX21 transcription start site (Supplemental Figure 17B). Collectively, informatic insights suggested that EBV-encoded transcriptional cotransactivators may mediate TBX21/T-bet expression and thereby affect ABC functions and responses. We also examined expression of cellular genes previously identified as biomarkers of EBV lytic reactivation (114) (Supplemental Figure 18). With few exceptions, biomarkers of the EBV lytic phase were strongly and broadly expressed in ABC 2. Collectively, these data suggest indirect roles of EBV infection in potentiating ABC pathogenicity. Without excluding other possible mechanisms, virus-related ABC responses may contribute to eMS and MS disease activity via inflammatory cytokine production.
Validation of ABC signature by flow cytometry of expanded eMS patient cohort. An expanded surface biomarker panel was applied to t2 PBMC samples from 18 participants in the original STAyCIS Trial (n = 7 LTNA, n = 11 SMSA outcomes) and healthy donors (n = 6). In addition to ABC markers and CXCR3, we evaluated PD-L1 (CD274) and PD-L2 (CD273) expression, since these genes were induced by EBV at the mRNA level and expressed in eMS ABCs from scRNA-Seq (Figure 5 and Supplemental Figure 19). The CD21lo fraction of the B cell compartment was significantly expanded in people with eMS who experienced subsequent MS activity versus healthy controls (1-sided Wilcoxon P = 0.031). CXCR3+ and CD11c+ B cell subsets were likewise enriched in eMS→SMSA outcomes. Similar results were observed for comparisons of SMSA to grouped healthy and LTNA samples (Figure 5, A–C). Moreover, eMS cohort samples contained significantly more CD21loCXCR3+ B cells (4.2% ± 3.9% versus 1.4% ± 0.7%, Wilcoxon P = 0.0091) and CD21loCXCR3+CD11c+ B cells (0.58% ± 0.63% versus 0.10% ± 0.06%, Wilcoxon P = 0.0047) than healthy controls. CD21loCXCR3+ B cells and CD21loCXCR3+CD11c+ B cells were likewise significantly enriched in eMS→SMSA outcomes versus healthy controls and all non-MS outcomes (healthy controls plus eMS→LTNA outcomes, n = 13), though differences between eMS→LTNA and eMS→SMSA outcomes were not significant (Figure 5, D and E). Thus, MS activity following initial presentation was associated not only with ABC enrichment specifically but also with broader enrichment of CXCR3+ and CD21lo cells across peripheral B cell niches. We also found that CXCR3+ ABCs (CD19+CD21loCD11c+) coexpressing the immune checkpoint ligands PD-L1 and PD-L2 were significantly enriched in people with eMS versus healthy controls (Wilcoxon P = 0.031; Figure 5F). Specifically, CD19+CD21loCXCR3+CD11c+ cells from 8 of 18 eMS samples contained a PD-L1+PD-L2+ subset compared with 0 of 6 healthy controls (Figure 5, G and H). Comparison of sex-matched samples identified enrichment of CD21loCXCR3+CD11c+ B cells from eMS→SMSA (n = 8) versus healthy controls (n = 6; Wilcoxon test P = 0.0013) and eMS→LTNA (n = 4; Wilcoxon P = 0.15) in female donors (Figure 5I). Similar ABC enrichment was observed in age-matched cohorts (36 to 45 years; eMS→SMSA n = 5; eMS→LTNA n = 6) (Figure 5J). Case-matched samples (n = 4 individuals) from eMS baseline and 6-month follow-up longitudinally exhibited elevated CD11c+, CXCR3+, and PD-L1+PD-L2+ subsets of CD21lo B cells, though these differences were not statistically significant (Supplemental Figure 19, B and C).
 Figure 5
Figure 5Flow cytometry validation of peripheral ABC expansion and immune checkpoint expression in people with eMS. (A) Quantification and statistical comparison of CD19+CD21lo cells among healthy (n = 6), all eMS (n = 18), eMS→LTNA (n = 7), eMS→SMSA (n = 11), and healthy + LTNA (n = 13) groups. All statistics represent P values from 1-sided Wilcoxon rank-sum tests. (B) Quantification and statistical comparison of CD19+CXCR3+ cells between groups as shown in A. (C) Quantification and statistical comparison of CD19+CD11c+ cells between groups as shown in A and B. (D) Quantification and statistical comparison of CD19+CD21loCXCR3+ cells between groups as shown in A–C. (E) Quantification and statistical comparison of ABCs (CD19+CD21loCXCR3+CD11c+) cells between groups as shown in A–D. (F) Quantification and statistical comparison of PD-L1+PD-L2+ ABCs (CD19+CD21loCXCR3+CD11c+) between groups as shown in A–E. (G) Summary of eMS and healthy samples exhibiting any PD-L1+PD-L2+ expression in CXCR3+ ABCs (CD19+CD21loCD11c+). The median frequency of such cells in positive cases is approximately 1 per 10,000 total B cells. (H) PD-L1 (CD274) and PD-L2 (CD273) coexpression in CD19+CD21loCXCR3+CD11c+ cells from select eMS→LTNA and eMS→SMSA but not healthy donors. (I) Sex-matched (female-only) quantification and statistical comparisons of CD19+CD21loCXCR3+CD11c+ cells among healthy (n = 6), eMS→LTNA (n = 4), and eMS→SMSA (n = 8) groups. Statistics represent P values from 2-sided Wilcoxon rank-sum tests. (J) Age-matched (36- to 45-year-olds) quantification and statistical comparisons of CD19+CD21loCXCR3+CD11c+ cells among healthy (n = 6), eMS→LTNA (n = 6), and eMS→SMSA (n = 5) groups. Statistics represent P values from 2-sided Wilcoxon rank-sum tests.
Collectively, flow cytometry validation experiments verified that ABCs are peripherally expanded in eMS — particularly in cases of additional MS activity — relative to healthy donors. These experiments further supported ABC upregulation of certain lineage-ectopic genes and critical immune checkpoint molecules associated with responses to EBV in a subset of eMS cases. Elevated PD-L1 in eMS versus healthy ABCs is particularly noteworthy given the recent identification of EBV+ PD-L1 cells in postmortem chronic active MS lesion tissue (115). However, we cannot definitively determine whether ABC responses in eMS are strictly dependent on direct EBV infection or reflect bystander effects based on our data.
-
Discussion
In summary, this study provides single-cell transcriptomic and protein-level evidence of ABCs with pathogenic features in people with eMS. These cells, which constituted about 5% of peripheral B cells, share many characteristics with ABCs in other disease contexts. Notably, eMS ABCs were distinguished from ABCs in other diseases by transcriptomic similarities to ABCs in response to EBV infection, which were identified in a prior in vitro study (34). Although EBV reads were not directly detected in eMS ABCs, we further found that the EBV-associated ABC gene signature was enriched in people with eMS who experienced additional disease activity versus individuals who did not during the STAyCIS Trial. A smaller subset of CD19+CD20+TBX21+ITGAX+ cells (ABC 2) from people with eMS (~0.3% of B cells) that exhibited surprisingly broad immune lineage expression and pro-inflammatory cytokine transcripts was also underrepresented in people without subsequent disease activity after baseline.
Prior studies have separately demonstrated EBV-induced CXCR3 expression in B cells (48), positive correlation between CXCR3+ B cell frequencies and EBV viral load in people with MS (64), and T-bet+CXCR3+ B cell induction and neuroinvasion in people with MS (50). In conjunction with these findings, the present identification of a shared gene signature — including CXCR3 — in EBV+ ABCs and ABCs from people with eMS absent in healthy controls further implies a link between neuroinvasive CXCR3+ ABCs and EBV infection. However, it is unclear whether such association may be direct or indirect, and definitive causality cannot be established in the absence of consistent viral detection. The enrichment of neuronal genes including cell adhesion molecules in the EBV-associated ABC signature is consistent with an acquired neurotropic capacity that may facilitate CNS infiltration as well as localized viral antigen presentation and inflammatory responses that precipitate MS. Because ABCs differentiate into PBs in response to innate and inflammatory stimuli (9), it is further conceivable that EBV infection could promote low-affinity antibody production by activated ABCs in the CNS. The likelihood of this pathogenic sequence of events would partially depend on the frequency of ABCs, which increases with age and is elevated in genetic females (1). In summary, this model is consistent with epidemiological aspects of MS, including age at time of EBV seroconversion and age- and sex-dependent accumulation of ABCs. Likewise, the efficacy of anti-CD20 therapy (116) and clinical exacerbations observed upon IFN-γ treatment (117) are each consistent with pathogenicity of CD20+ ABCs, which proliferate and differentiate in response to IFN-γ. Notably, this model also aligns with empirical findings and prevailing hypotheses (55, 118, 119) of EBV etiology in MS: molecular mimicry (56), pro-inflammatory bystander damage (94, 103), and the potential for antibody production by autoreactive clones, the latter of which is a frequent fate of extrafollicular activated ABCs (12, 13).
Detection of EBV genomes, transcripts, and proteins has been reported in MS brain lesions; however, negative results in multiple studies — in some cases using the same samples — precludes definitive functional insights linking viral antigens to MS pathology (58–63). The same is true regarding the lack of detected EBV reads within previous findings from an scRNA-Seq study of people with MS (49). Similarly, the present study did not directly detect EBV in B cells from early MS clinical samples. This may be due to one or more factors, including transcriptome undersampling inherent to current scRNA-Seq methods (120); inefficient capture chemistry for the most abundant EBV transcripts, which are not polyadenylated (121); limited viral gene expression owing to restricted latency programs or failure to establish successful infection (34, 122–124); or a combination thereof. In this case, the transcriptomic profile exhibited by ABCs in eMS and in response to EBV infection in vitro may reflect epigenetically encoded responses of the ABC niche that EBV, a prevalent stimulus, is capable of eliciting. Likewise, although our prior work demonstrated that EBV can infect ABCs based on viral read detection (34), we cannot definitively rule out the possibility that EBV-induced responses of other infected cells (e.g., inflammatory cytokine production) might indirectly activate bystander ABCs that do not harbor the virus.
There are several limitations to this study. Primarily, our findings do not definitively identify EBV mechanistic involvement in early MS. Specifically, there is a current lack of insight with respect to (a) whether the observed age dependence of EBV seroconversion for risk in MS is directly related to the increase in ABC frequency over time; (b) whether EBV promotes or otherwise enhances the accumulation of ABCs in vivo; and (c) whether periodic EBV reactivation can result in chronic de novo infection of ABCs or other B cell subtypes, consistent with autoimmune disease flares. The number of LTNA samples analyzed with scRNA-Seq was small; however, flow cytometric validation of the ABC enrichment and checkpoint molecule expression in a larger number of LTNA cases provides additional support for the observed results. Also, this study did not examine patient- and time point–matched CSF samples because of limited availability. Future efforts to contextualize ABC expansion and EBV biomarkers in peripheral blood with biomarker signatures in CSF will be highly valuable. Additional studies are required to investigate the consequences of EBV infection in different genetic backgrounds with known autoimmune susceptibility variants. Thus, the nature of EBV infection in promoting the formation and activation of ABCs and the relevance of this host-virus relationship in MS and other autoimmune diseases warrants substantial focus in future work. As noted previously, samples analyzed herein were obtained from individuals diagnosed with CIS using the 2001 McDonald criteria, many of whom would likely have been diagnosed with MS at baseline using current standards. Thus, future studies should examine the associations and pathologic involvement of ABCs on a larger scale in clinical cohorts of people with eMS using updated diagnostic criteria.
Despite these unknowns, the present work advances our understanding of alterations to the peripheral B cell landscape in patients during the early stages of MS, biomarkers of peripheral B cells to distinguish differential disease activity in MS, and evidence of pro-inflammatory ABC expansion and immune checkpoint evasion. Likewise, the data herein underscore the value and importance of addressing questions of viral pathogenesis related to disease at cellular resolution with high-dimensional techniques.
-
Methods
Sex as a biological variable. Our gene expression study included 32 human PBMC samples from 16 individuals. As expected given the distribution of the disease under study, these were split as 11 females and 5 males. More demographic details regarding the trial and this specific cohort are noted below.
Statistics. Statistical comparisons within this study were made as described above and as denoted in the text. Broadly, genes found to be differentially expressed in this study were identified based on significance after multiple-hypothesis testing (Bonferroni’s correction) using the Seurat FindMarkers function. Further, statistical significance of differential expression levels for select genes of interest expression were calculated via 2-sided Wilcoxon rank-sum test. Significant differences in gene expression distributions were determined via 2-sided KS test. Significant differences in cell phenotype frequencies (measured by scRNA-Seq, CITE-Seq, and flow cytometry) between groups were determined using Wilcoxon rank-sum tests. P < 0.05 was considered statistically significant.
Study approval. The original STAyCIS Trial received institutional review board (IRB) approval from 14 centers in the United States and Canada. Single-cell analyses of samples from the STAyCIS Trial were additionally approved by a Duke University IRB. All participants provided written informed consent prior to study entry.
Data availability. All clinical data associated with the STAyCIS Trial are publicly available on the TrialShare website, https://www.itntrialshare.org/project/Studies/ITN020AIPUBLIC/Study%20Data/begin.view B cell scRNA-Seq + CITE-Seq data from the eMS cohort reported herein are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession GSE267750). Additional methods are available in Supplemental Methods.
-
Author contributions
EDS, SGG, and MAL conceived and designed the study and wrote the manuscript. E Haukenfrers, VJ, JG, KA, and E Hocke performed single-cell experiments and curated public datasets. EDS and VJ processed the data and extracted B cell populations. EDS analyzed the sequencing data. EDS and GQH performed flow cytometry and RNA Flow-FISH experiments and analyses. LAC, KMH, and SSZ reviewed and edited the manuscript. All authors have approved the final version of the manuscript.
-
Acknowledgments
The authors wish to thank Jay Tourigny (Duke University) for assistance with cytokine secretion-profiling experiments reported herein and the ITN for thoughtful feedback in the development of this manuscript. We also thank Nicolas Reinoso in the Luftig laboratory for thoughtful discussion and feedback. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH; award UM1AI109565) and an ITN award to SGG. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. EDS wishes to acknowledge funding from the American Cancer Society (award PF-21-084-01-DMC). MAL acknowledges support from the NIH National Institute of Dental and Craniofacial Research (R01DE025994). GQH was supported by the Duke Basic Immunology Training Program (T32AI052077).
Address correspondence to: Elliott D. SoRelle, Medical Sciences II, Room 6748A, 1137 Catherine Street, Ann Arbor, Michigan, 48109, USA. Phone: 734.763.3531; Email: sorelle@med.umich.edu. Or to: Simon G. Gregory, Carmichael Building 51-102, 300 N. Duke Street, Durham, North Carolina, 27701, USA. Phone: 919.684.0726; Email: simon.gregory@duke.edu. Or to: Micah A. Luftig, CARL Building 0036, 213 Research Drive, Durham, North Carolina, 27710, USA. Phone: 919.668.3091; Email: micah.luftig@duke.edu.
-
Footnotes
EDS’s present address is: Department of Microbiology & Immunology, University of Michigan Medical School, Ann Arbor, Michigan, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2025, SoRelle et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2025;10(12):e188543.https://doi.org/10.1172/jci.insight.188543.
-
References
- Rubtsov AV, et al. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c+ B-cell population is important for the development of autoimmunity. Blood. 2011;118(5):1305–1315.
- Ehrhardt GR, et al. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med. 2008;205(8):1807–1817.
- Rubtsova K, et al. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc Natl Acad Sci U S A. 2013;110(34):E3216–E3224.
- Rubtsova K, et al. Age-associated B cells: a T-bet-dependent effector with roles in protective and pathogenic immunity. J Immunol. 2015;195(5):1933–1937.
- Sohn HW, et al. FcRL4 acts as an adaptive to innate molecular switch dampening BCR signaling and enhancing TLR signaling. Blood. 2011;118(24):6332–6341.
- Naradikian MS, et al. Cutting edge: IL-4, IL-21, and IFN-γ interact to govern T-bet and CD11c expression in TLR-activated B Cells. J Immunol. 2016;197(4):1023–1028.
- Portugal S, et al. Atypical memory B cells in human chronic infectious diseases: an interim report. Cell Immunol. 2017;321:18–25.
- Rubtsova K, et al. TLR7, IFNγ, and T-bet: their roles in the development of ABCs in female-biased autoimmunity. Cell Immunol. 2015;294(2):80–83.
- Nicholas MW, et al. A novel subset of memory B cells is enriched in autoreactivity and correlates with adverse outcomes in SLE. Clin Immunol. 2008;126(2):189–201.
- Isnardi I, et al. Complement receptor 2/CD21- human naive B cells contain mostly autoreactive unresponsive clones. Blood. 2010;115(24):5026–5036.
- Souyris M, et al. TLR7 escapes X chromosome inactivation in immune cells. Sci Immunol. 2018;3(19):eaap8855.
- Naradikian MS, et al. Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunol Rev. 2016;269(1):118–129.
- Myles A, et al. T-bet+ B cells: a common denominator in protective and autoreactive antibody responses? Curr Opin Immunol. 2019;57:40–45.
- Johnson JL, et al. The transcription factor T-bet resolves memory B cell subsets with distinct tissue distributions and antibody specificities in mice and humans. Immunity. 2020;52(5):842–855.
- Karnell JL, et al. Role of CD11c+ T-bet+ B cells in human health and disease. Cell Immunol. 2017;321:40–45.
- Knox JJ, et al. T-bet+ memory B cells: generation, function, and fate. Immunol Rev. 2019;288(1):149–160.
- Rubtsova K, et al. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J Clin Invest. 2017;127(4):1392–1404.
- Wang S, et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat Commun. 2018;9(1):1758.
- Scharer CD, et al. Epigenetic programming underpins B cell dysfunction in human SLE. Nat Immunol. 2019;20(8):1071–1082.
- Verstappen GM, et al. Gene expression profiling of epithelium-associated FcRL4+ B cells in primary Sjögren’s syndrome reveals a pathogenic signature. J Autoimmun. 2020;109:102439.
- Tsubaki T, et al. Accumulation of plasma cells expressing CXCR3 in the synovial sublining regions of early rheumatoid arthritis in association with production of Mig/CXCL9 by synovial fibroblasts. Clin Exp Immunol. 2005;141(2):363–371.
- Yeo L, et al. Expression of FcRL4 defines a pro-inflammatory, RANKL-producing B cell subset in rheumatoid arthritis. Ann Rheum Dis. 2015;74(5):928–935.
- Orton S-M, et al. Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 2006;5(11):932–936.
- Weiss GE, et al. Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. J Immunol. 2009;183(3):2176–2182.
- Ho J, et al. Two overrepresented B cell populations in HIV-infected individuals undergo apoptosis by different mechanisms. Proc Natl Acad Sci U S A. 2006;103(51):19436–19441.
- Moir S, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008;205(8):1797–1805.
- Holla P, et al. Shared transcriptional profiles of atypical B cells suggest common drivers of expansion and function in malaria, HIV, and autoimmunity. Sci Adv. 2021;7(22):eabg8384.
- Austin JW, et al. Overexpression of T-bet in HIV infection is associated with accumulation of B cells outside germinal centers and poor affinity maturation. Sci Transl Med. 2019;11(520):eaax0904.
- Hopp CS, et al. Atypical B cells up-regulate costimulatory molecules during malaria and secrete antibodies with T follicular helper cell support. Sci Immunol. 2022;7(71):eabn1250.
- Gewurz B, et al. Epstein-Barr virus. In: Howley PM, Knipe DM, eds. Fields Virology. Wolters Kluwer; 2021:324–388.
- Roughan JE, Thorley-Lawson DA. The intersection of Epstein-Barr virus with the germinal center. J Virol. 2009;83(8):3968–3976.
- Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol. 2001;1(1):75–82.
- Babcock GJD L. EBV persistence in memory B cells in vivo. Immunity. 1998;9(3):395–404.
- SoRelle ED, et al. Time-resolved transcriptomes reveal diverse B cell fate trajectories in the early response to Epstein-Barr virus infection. Cell Rep. 2022;40(9):111286.
- Cohen JI, et al. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med. 2011;3(107):107fs7.
- Ascherio A, Munger KL. EBV and autoimmunity. In: Münz C, ed. Epstein Barr Virus Volume 1. Springer Cham; 2015:365–385.
- Gross AJ, et al. EBV and systemic lupus erythematosus: a new perspective. J Immunol. 2005;174(11):6599–6607.
- Tosato G, et al. Defective EBV-specific suppressor T-cell function in rheumatoid arthritis. N Engl J Med. 1981;305(21):1238–1243.
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–768.
- Fox R, et al. Detection of Epstein-Barr virus-associated antigens and DNA in salivary gland biopsies from patients with Sjogren’s syndrome. J Immunol. 1986;137(10):3162–3168.
- Saito I, et al. Detection of Epstein-Barr virus DNA by polymerase chain reaction in blood and tissue biopsies from patients with Sjogren’s syndrome. J Exp Med. 1989;169(6):2191–2198.
- Tosato G, et al. Abnormally elevated frequency of Epstein-Barr virus-infected B cells in the blood of patients with rheumatoid arthritis. J Clin Invest. 1984;73(6):1789–1795.
- Balandraud N, et al. Epstein-Barr virus and rheumatoid arthritis. Autoimmun Rev. 2004;3(5):362–367.
- Tsokos G, et al. Epstein-Barr virus induces normal B cell responses but defective suppressor T cell responses in patients with systemic lupus erythematosus. J Immunol. 1983;131(4):1797–1801.
- Kang I, et al. Defective control of latent Epstein-Barr virus infection in systemic lupus erythematosus. J Immunol. 2004;172(2):1287–1294.
- Poole BD, et al. Aberrant Epstein-Barr viral infection in systemic lupus erythematosus. Autoimmun Rev. 2009;8(4):337–342.
- Larsen M, et al. Exhausted cytotoxic control of Epstein-Barr virus in human lupus. PLoS Pathog. 2011;7(10):e1002328.
- SoRelle ED, et al. Epstein-Barr virus perpetuates B cell germinal center dynamics and generation of autoimmune-associated phenotypes in vitro. Front Immunol. 2022;13:1001145.
- Ramesh A, et al. A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis. Proc Natl Acad Sci U S A. 2020;117(37):22932–22943.
- van Langelaar J, et al. Induction of brain-infiltrating T-bet-expressing B cells in multiple sclerosis. Ann Neurol. 2019;86(2):264–278.
- Ascherio A, et al. Epstein-Barr virus antibodies and risk of multiple sclerosis: a prospective study. JAMA. 2001;286(24):3083–3088.
- Bjornevik K, et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. 2022;375(6578):296–301.
- Bar-Or A, et al. Epstein-Barr Virus in Multiple Sclerosis: theory and emerging immunotherapies. Trends Mol Med. 2020;26(3):296–310.
- Pender MP. Infection of autoreactive B lymphocytes with EBV, causing chronic autoimmune diseases. Trends Immunol. 2003;24(11):584–588.
- Soldan SS, Lieberman PM. Epstein–Barr virus and multiple sclerosis. Nat Rev Microbiol. 2023;21(1):51–64.
- Lanz TV, et al. Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM. Nature. 2022;603(7900):321–327.
- Lünemann JD, et al. Increased frequency and broadened specificity of latent EBV nuclear antigen-1-specific T cells in multiple sclerosis. Brain. 2006;129(pt 6):1493–1506.
- Serafini B, et al. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14(2):164–174.
- Serafini B, et al. Dysregulated Epstein-Barr virus infection in the multiple sclerosis brain. J Exp Med. 2007;204(12):2899–2912.
- Lassmann H, et al. Epstein-Barr virus in the multiple sclerosis brain: a controversial issue--report on a focused workshop held in the Centre for Brain Research of the Medical University of Vienna, Austria. Brain. 2011;134(pt 9):2772–2786.
- Moreno MA, et al. Molecular signature of Epstein-Barr virus infection in MS brain lesions. Neurol Neuroimmunol Neuroinflamm. 2018;5(4):e466.
- Magliozzi R, et al. B-cell enrichment and Epstein-Barr virus infection in inflammatory cortical lesions in secondary progressive multiple sclerosis. J Neuropathol Exp Neurol. 2013;72(1):29–41.
- Tzartos JS, et al. Association of innate immune activation with latent Epstein-Barr virus in active MS lesions. Neurology. 2012;78(1):15–23.
- van Langelaar J, et al. The association of Epstein-Barr virus infection with CXCR3+ B-cell development in multiple sclerosis: impact of immunotherapies. Eur J Immunol. 2021;51(3):626–633.
- Waubant E, et al. Randomized controlled trial of atorvastatin in clinically isolated syndrome: the STAyCIS study. Neurology. 2012;78(15):1171–1178.
- Jacobs LD, et al. Intramuscular interferon beta-1a therapy initiated during a first demyelinating event in multiple sclerosis. CHAMPS Study Group. N Engl J Med. 2000;343(13):898–904.
- McDonald WI, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50(1):121–127.
- Miller DH, et al. Clinically isolated syndromes. Lancet Neurol. 2012;11(2):157–169.
- O’riordan J, et al. The prognostic value of brain MRI in clinically isolated syndromes of the CNS. A 10-year follow-up. Brain. 1998;121 (Pt 3) (3):495–503.
- Montalban X, et al. MRI criteria for MS in patients with clinically isolated syndromes. Neurology. 2010;74(5):427–434.
- Kuhle J, et al. Conversion from clinically isolated syndrome to multiple sclerosis: A large multicentre study. Mult Scler. 2015;21(8):1013–1024.
- Schafflick D, et al. Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat Commun. 2020;11(1):247.
- Nehar-Belaid D, et al. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat Immunol. 2020;21(9):1094–1106.
- Hong X, et al. Single-cell RNA sequencing reveals the expansion of cytotoxic CD4+ T lymphocytes and a landscape of immune cells in primary Sjögren’s syndrome. Front Immunol. 2021;11:594658.
- Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.
- Sutton HJ, et al. Atypical B cells are part of an alternative lineage of B cells that participates in responses to vaccination and infection in humans. Cell Rep. 2021;34(6):108684.
- Yang R, et al. Human T-bet governs the generation of a distinct subset of CD11chighCD21low B cells. Sci Immunol. 2022;7(73):eabq3277.
- Keller B, et al. The expansion of human T-bethighCD21low B cells is T cell dependent. Sci Immunol. 2021;6(64):eabh0891.
- Knutson JC. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J Virol. 1990;64(6):2530–2536.
- Cheah MS, et al. fgr proto-oncogene mRNA induced in B lymphocytes by Epstein-Barr virus infection. Nature. 1986;319(6050):238–240.
- Martin HJ, et al. Manipulation of the toll-like receptor 7 signaling pathway by Epstein-Barr virus. J Virol. 2007;81(18):9748–9758.
- Ambegaonkar AA, et al. Atypical B cells in chronic infectious diseases and systemic autoimmunity: puzzles with many missing pieces. Curr Opin Immunol. 2022;77:102227.
- Looijenga LH, et al. POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res. 2003;63(9):2244–2250.View this article via: PubMed Google Scholar
- Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391.
- Calvanese V, et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature. 2019;576(7786):281–286.
- Vercauteren SM, Sutherland HJ. Constitutively active Notch4 promotes early human hematopoietic progenitor cell maintenance while inhibiting differentiation and causes lymphoid abnormalities in vivo. Blood. 2004;104(8):2315–2322.
- Dehmel T, et al. The cell-specific expression of metalloproteinase-disintegrins (ADAMs) in inflammatory myopathies. Neurobiol Dis. 2007;25(3):665–674.
- Qin S, et al. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest. 1998;101(4):746–754.
- Mohan K, et al. CXCR3 is required for migration to dermal inflammation by normal and in vivo activated T cells: differential requirements by CD4 and CD8 memory subsets. Eur J Immunol. 2005;35(6):1702–1711.
- Xu J, et al. Chemokine receptor CXCR3 is required for lethal brain pathology but not pathogen clearance during cryptococcal meningoencephalitis. Sci Adv. 2020;6(25):eaba2502.
- Marques CP, et al. CXCR3-dependent plasma blast migration to the central nervous system during viral encephalomyelitis. J Virol. 2011;85(13):6136–6147.
- Teng Y, et al. The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT. 2014;3(1):e28086.
- Teng Y, et al. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer. 2010;103(7):1066–1075.
- Bar-Or A, et al. Abnormal B-cell cytokine responses a trigger of T-cell-mediated disease in MS? Ann Neurol. 2010;67(4):452–461.
- Karni A, et al. IL-18 is linked to raised IFN-gamma in multiple sclerosis and is induced by activated CD4(+) T cells via CD40-CD40 ligand interactions. J Neuroimmunol. 2002;125(1–2):134–140.
- Bartosik-Psujek H, Stelmasiak Z. The levels of chemokines CXCL8, CCL2 and CCL5 in multiple sclerosis patients are linked to the activity of the disease. Eur J Neurol. 2005;12(1):49–54.
- Zaiss DM, et al. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42(2):216–226.
- Yang P, et al. Role of PDGF-D and PDGFR-β in neuroinflammation in experimental ICH mice model. Exp Neurol. 2016;283(pt a):157–164.
- De Felice B, et al. Small non-coding RNA signature in multiple sclerosis patients after treatment with interferon-β. BMC Med Genomics. 2014;7(1):26.
- Zhao Y, et al. The RNA quality control pathway nonsense-mediated mRNA decay targets cellular and viral RNAs to restrict KSHV. Nat Commun. 2020;11(1):3345.
- Abernathy E, Glaunsinger B. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology. 2015;479–480:600–608.
- Rigby RE, Rehwinkel J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 2015;36(3):179–188.
- Li R, et al. Proinflammatory GM-CSF-producing B cells in multiple sclerosis and B cell depletion therapy. Sci Transl Med. 2015;7(310):310ra166.
- Rauch PJ, et al. Innate response activator B cells protect against microbial sepsis. Science. 2012;335(6068):597–601.
- Iparraguirre L, et al. Whole-transcriptome analysis in peripheral blood mononuclear cells from patients with lipid-specific oligoclonal IgM band characterization reveals two circular RNAs and two linear RNAs as biomarkers of highly active disease. Biomedicines. 2020;8(12):540.
- Speir M, Lawlor KE. RIP-roaring inflammation: RIPK1 and RIPK3 driven NLRP3 inflammasome activation and autoinflammatory disease. Semin Cell Dev Biol. 2021;109:114–124.
- Lawlor KE, et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat Commun. 2015;6(1):6282.
- Jiang Y, et al. The protective role of NR4A3 in acute myocardial infarction by suppressing inflammatory responses via JAK2-STAT3/NF-κB pathway. Biochem Biophys Res Commun. 2019;517(4):697–702.
- Freire PR, Conneely OM. NR4A1 and NR4A3 restrict HSC proliferation via reciprocal regulation of C/EBPα and inflammatory signaling. Blood. 2018;131(10):1081–1093.
- Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471.
- Shi C-S, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13(3):255–263.
- Anand PK, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488(7411):389–393.
- McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501.
- Arvey A, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12(2):233–245.
- Serafini B, et al. EBV infected cells in the multiple sclerosis brain express PD-L1: How the virus and its niche may escape immune surveillance. J Neuroimmunol. 2024;389:578314.
- Hauser SL, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358(7):676–688.
- Panitch H, et al. Exacerbations of multiple sclerosis in patients treated with gamma interferon. Lancet. 1987;1(8538):893–895.
- Fujiwara S, Takei M. Are viral infections key inducers of autoimmune diseases? Focus on Epstein-Barr virus. Viruses. 2015;14(9):38.
- Pender MP, Burrows SR. Epstein-Barr virus and multiple sclerosis: potential opportunities for immunotherapy. Clin Transl Immunology. 2014;3(10):e27.
- Streets AM, Huang Y. How deep is enough in single-cell RNA-seq? Nat Biotechnol. 2014;32(10):1005–1006.
- Lerner MR, et al. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981;78(2):805–809.
- McFadden K, et al. Metabolic stress is a barrier to Epstein-Barr virus-mediated B-cell immortalization. Proc Natl Acad Sci U S A. 2016;113(6):E782–E790.
- Price AM, Luftig MA. To be or not IIb: a multi-step process for Epstein-Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 2015;11(3):e1004656.
- Nikitin PA, et al. Mitogen-induced B-cell proliferation activates Chk2-dependent G1/S cell cycle arrest. PLoS One. 2014;9(1):e87299.
- Alquicira-Hernandez J, Powell JE. Nebulosa recovers single-cell gene expression signals by kernel density estimation. Bioinformatics. 2021;37(16):2485–2487.
-
Version history
- Version 1 (May 13, 2025): In-Press Preview
- Version 2 (June 23, 2025): Electronic publication









