Research ArticleImmunologyOncology Free access | 10.1172/jci.insight.130000
Resident memory CD8+ T cells within cancer islands mediate survival in breast cancer patients
Colt A. Egelston,1 Christian Avalos,1 Travis Y. Tu,1 Anthony Rosario,1 Roger Wang,1 Shawn Solomon,1 Gayathri Srinivasan,1 Michael S. Nelson,2 Yinghui Huang,1 Min Hui Lim,1 Diana L. Simons,1 Ting-Fang He,1 John H. Yim,3 Laura Kruper,3 Joanne Mortimer,4 Susan Yost,4 Weihua Guo,1 Christopher Ruel,5 Paul H. Frankel,5 Yuan Yuan,4 and Peter P. Lee1
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by
Egelston, C.
in:
PubMed
|
Google Scholar
|

1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by
Avalos, C.
in:
PubMed
|
Google Scholar
|

1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by
Tu, T.
in:
PubMed
|
Google Scholar
|

1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Rosario, A. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Wang, R. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Solomon, S. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Srinivasan, G. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by
Nelson, M.
in:
PubMed
|
Google Scholar
|

1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Huang, Y. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Lim, M. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Simons, D. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by He, T. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Yim, J. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Kruper, L. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Mortimer, J. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Yost, S. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by
Guo, W.
in:
PubMed
|
Google Scholar
|

1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Ruel, C. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Frankel, P. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Yuan, Y. in: PubMed | Google Scholar
1Department of Immuno-Oncology,
2Light Microscopy and Digital Imaging Core,
3Department of Surgery,
4Department of Medical Oncology, and
5Department of Biostatistics, Beckman Research Institute, City of Hope, Duarte, California, USA.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
Find articles by Lee, P. in: PubMed | Google Scholar
Published August 29, 2019 - More info
JCI Insight. 2019;4(19):e130000. https://doi.org/10.1172/jci.insight.130000.
© 2019 American Society for Clinical Investigation
Received: May 2, 2019; Accepted: August 23, 2019
-
Abstract
CD8+ tumor-infiltrating lymphocytes (TILs) correlate with relapse-free survival (RFS) in most cancer types, including breast cancer. However, subset composition, functional status, and spatial location of CD8+ TILs in relation to RFS in human breast tumors remain unclear. Spatial tissue analysis via quantitative immunofluorescence showed that infiltration of CD8+ T cells into cancer islands was more significantly associated with RFS than CD8+ T cell infiltration into either tumor stroma or total tumor. Localization into cancer islands within tumors is mediated by expression of the integrin CD103, which is a marker for tissue-resident memory T cells (TRMs). Analysis of fresh tumor samples revealed that CD8+ TRMs are functionally similar to other CD8+ TILs, suggesting that the basis of their protective effect is their spatial distribution rather than functional differences. Indeed, CD103+ TRMs, as compared with CD103–CD8+ TILs, are enriched within cancer islands, and CD8+ TRM proximity to cancer cells drives the association of CD8+ TIL densities with RFS. Together, these findings reveal the importance of cancer island–localized CD8+ TRMs in surveillance of the breast tumor microenvironment and as a critical determinant of RFS in patients with breast cancer.
-
Introduction
The presence of tumor-infiltrating lymphocytes (TILs) associates favorably with relapse-free survival (RFS) in breast cancer (1–4). CD8+ TILs in human breast tumors have been demonstrated to be primarily antigen-experienced T cells, but little else is known about the relationship between T cell composition and spatial localization within the tumor microenvironment with RFS (5). Successful immunotherapy of breast cancer necessitates a greater understanding of the T cell infiltrate in breast tumors.
Breast tumors are segregated by receptor expression patterns into 3 major subtypes: estrogen receptor expressing, human epidermal growth factor receptor 2 (HER2) enriched, and basal-like/triple negative (6). These subtypes correlate with more detailed molecular expression subtypes and differ in therapeutic response and prognosis (7). Triple-negative breast cancer (TNBC) is an aggressive breast cancer subset with a higher frequency of early relapse and significantly shorter time to recurrence as compared with estrogen receptor (ER+) breast cancer (8). Among breast cancer subtypes, TNBC and HER2+ have been clearly defined to have a positive association between RFS and presence of tumor-infiltrating T cells (4).
Tumors may also be divided by their immune infiltration profiles: immune inflamed, immune excluded, and immune desert (9). Immune-inflamed tumors involve infiltration of T cells into and around the tumor parenchyma (cancer islands), while immune-excluded tumors have T cell infiltration only into tumor stroma but not cancer islands. Immune-desert tumors lack T cell infiltration altogether, either in cancer islands or stroma. Beyond patient prognosis, higher levels of T cell infiltration in tumors have been shown to correlate positively with response rates to immunotherapies (10, 11). Thus, analysis of TIL infiltration patterns and characteristics is valuable for mechanistic and clinical insights into patient outcomes.
CD103, an αE integrin, is a key marker for tissue-resident memory T cells (TRMs), with a functional role in retaining T cells within peripheral tissues via binding to E-cadherin on epithelial cells (12). In addition to CD103, another key marker for CD8+ TRMs is CD69, which further limits T cell tissue egress by promoting downregulation of the chemotaxis receptor G protein–coupled sphingosine 1-phosphate receptor-1 (S1P1) (13). Together, CD69 and CD103 are crucial determinants of CD8+ TRMs’ retention and accumulation in epithelial cell–rich tissues, such as intestinal tissue, skin, lung respiratory tissue, and salivary glands (14–18).
CD8+ TRMs have been identified as key immune players in tumor microenvironments, with survival associations in ovarian cancer, non-small cell lung carcinoma, and melanoma (19–21). Tumor-infiltrating CD103+CD8+ TRMs have been implicated to be associated with improved survival prognosis in head and neck cancer, non–small cell lung cancer, ovarian cancer, and skin cancer patients (20–23). Tumor-associated TRMs from different malignancies have demonstrated varying degrees of checkpoint molecule expression profiles and heterogeneity in functional capacities (24). A clearer understanding of the role of CD8+ TRMs in the breast tumor microenvironment and their relationship with prognosis is needed.
Here, phenotypic analysis of fresh and archival breast tumors unraveled mechanisms of this association. Using quantitative spatial image analysis, we demonstrate that CD103+CD8+ TILs are enriched in the cancer islands of breast tumors. Functional profiling revealed similar cytokine production capacity of CD103+CD8+ TRMs as compared to CD103–CD8+ TILs, highlighting spatial localization of CD103+CD8+ TRMs as a key phenotypic difference of this TIL subset. Finally, we demonstrate that increased densities of CD103+CD8+ TILs in cancer islands within breast tumors is more significantly associated with RFS than CD8+ TILs within stroma. These results demonstrate that CD8+ TRMs are a major component of immune-inflamed breast tumors and play an important role in clinical outcome.
-
Results
CD103+CD8+ T cells localize to cancer islands in tumors and epithelial areas in noncancerous breast tissue. Because CD103 engagement with E-cadherin has been shown to mediate T cell retention in epithelial tissues, we hypothesized that CD103 expression by CD8+ T cells resulted in unique localization within breast tissues. Human breast tumors are composed of epithelial cell–derived “cancer islands” and interlaced “stromal” areas composed of fibroblasts, mesenchymal stem cells, and various immune cells. Using quantitative immunofluorescence (QIF), we were able to assess spatial localization of CD8+ T cells in tumor tissues within tissue-segmented classifications of either cancer islands (pan-cytokeratin–positive [CK+] areas) or stromal areas (CK– areas), as outlined in Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130000DS1). Similarly staged TNBC primary tumor formalin-fixed, paraffin-embedded (FFPE) tissues (Supplemental Table 1) from patients with at least 5 years of follow-up were interrogated using QIF. These patients had no therapy before surgery and underwent similar chemotherapy regimens following surgery, allowing us to interrogate the relationship between the immune tumor microenvironment and RFS in treatment-naive tumors.
Tumor tissues and noncancerous breast tissues (NCBTs) were costained for markers CK, CD8, and CD103 (Figure 1A). In breast tumors, CD8+ T cell density was significantly higher in stromal areas than in cancer islands (Figure 1B). However, CD103+CD8+ T cells were highly enriched within the cancer islands of tumor tissues (Figure 1C). Similarly, in NCBTs, nearly all CD8+ T cells in the epithelial ducts expressed CD103, while the majority of CD8+ T cells in stromal areas were CD103– (Figure 1D and additional representative images in Supplemental Figure 2). Together, these data show significant localization of CD103+CD8+ T cells to cancer islands in tumors and epithelial areas in general in human breast tissues.
 Figure 1
Figure 1CD103+CD8+ T cells localize to epithelial cell regions in breast tumors and non-cancerous breast tissues. (A) Formalin-fixed, paraffin-embedded (FFPE) tissues were assayed for expression of CD103 on CD8+ T cells by quantitative immunofluorescence (QIF). Whole-tissue sections from breast tumors or noncancerous breast tissues (NCBTs) were stained and imaged, followed by quantitative analysis of representative fields. Pan-cytokeratin (CK; shown in gray), CD8 (shown in cyan), and CD103 (shown in red) stains are depicted as composite images. Tissue segmentation algorithms based on CK staining allowed for distinct identification of cancer islands and stroma areas or epithelial ducts and stroma areas in tumors and NCBTs, respectively. T cell phenotypes (cyan dots, CD8+CD103–; red dots, CD8+CD103+) were created as shown for quantification and localization within segmented tissues. Scale bars: 50 μm. (B) Total CD8+ T cell density in either cancer islands or stroma was assessed. The percentages of CD8+ T cells expressing CD103 within segmented areas of breast tumors (C) and NCBTs (D) were assessed. Each symbol represents data from a unique patient sample. Tumor samples n = 25. NCBT samples n = 8. Significance was calculated using 2-tailed Student’s t tests. ****P < 0.0001.
CD103+ TRMs are a major component of CD8+ TILs in human breast tumors. Expression of both CD103 and CD69 has been tied to CD8+ TRM T cells’ localization and retention within peripheral tissues. To examine the phenotype of CD103+CD8+ T cells in human breast tissues, we obtained fresh surgically discarded breast tumors (both TNBC and ER+), NCBTs, and matched peripheral blood mononuclear cells (PBMCs) (Supplemental Tables 2 and 3). Single-cell suspensions of digested tissues were analyzed by flow cytometry for canonical markers of memory T cells (Figure 2, A–C, and gating strategy in Supplemental Figure 3). CD8+ T cells in both breast tumors and NCBTs were composed primarily of CD45RA–CCR7– effector memory cells. Further profiling of memory CD8+ T cells revealed that a large population coexpressed both CD69 and CD103 in breast tumors and NCBT, while CD69+CD103+CD8+ T cells were rarely found in the PBMCs of patients with breast cancer. Memory composition and frequencies of CD69+CD103+CD8+ T cells were similar in ER+ and TNBC tumors, identifying them as major cell populations in the tumor microenvironment of human breast tumors (Supplemental Figure 4, A and B).
 Figure 2
Figure 2CD8+ tissue-resident memory T cells are a major population of CD8+ T cells in human breast tumors and NCBTs. (A) Single-cell suspensions from peripheral blood mononuclear cells (PBMCs), tumors, and NCBTs were examined for expression of memory T cell and tissue-resident memory T cell (TRM) canonical markers CD45RA, CCR7, CD69, and CD103 by flow cytometry as shown. (B) Frequencies of CD8+ T cells in each tissue compartment that were CD45RA+CCR7+ (naive), CD45RA–CCR7+ (central memory, CM), CD45RA–CCR7– (effector memory, EM), or CD45RA+CCR7– (effector memory RA+, EMRA) are summarized. (C) Frequencies of CD45RA–CD8+ T cells in each tissue compartment expressing various patterns of CD69 and CD103 are summarized. (D) CD103+CD8+ T cells and CD103–CD8+ T cells from breast tumors and NCBTs were assessed by real-time PCR for gene expression. Gene expression and statistics shown are relative to control circulating memory CD8+ T cells. Each symbol represents data from a unique patient sample. Tumor samples n = 36. NCBT samples n = 21. PBMC samples n = 24. Significance was calculated using 1-way ANOVA and Holm-Šídák multiple-comparisons tests. *P < 0.05; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
A distinct TRM gene expression signature has previously been identified for CD8+ T cells, including upregulation of ITGAE, ITGA1, CD244, and XCL1 and downregulation of S1PR1 (25). We examined the RNA expression levels of these genes in CD103+ and CD103–CD8+ T cell populations from breast tumors and NCBTs relative to circulating memory CD8+ T cells (Figure 2D). As expected, RNA levels of ITGAE were significantly higher in CD103+CD8+ T cells relative to both circulating memory CD8+ T cells and CD103–CD8+ T cells. CD103+CD8+ T cells also had significantly lower expression of S1PR1 relative to both circulating memory CD8+ T cells and tissue CD103–CD8+ T cells, suggesting a lack of circulation reentry potential by these cells. Additionally, gene expression of ITGA1, CD244, and XCL1 was significantly higher in CD103+ T cells compared with circulating memory CD8+ T cells in both breast tumor tissue and NCBT, demonstrating them as bona fide TRMs. Interestingly, CD103–CD8+ T cells also showed decreased levels of S1PR1 and increased levels of ITGA1, CD244, and XCL1 in comparison with circulating memory CD8+ T cells, suggesting that they may be transitioning to a TRM phenotype as well. This is also reflected by the large fraction of CD103–CD8+ T cells in both breast tumors and NCBTs expressing CD69 (Figure 2, A and C), a molecule that plays an important role in inhibition of S1PR1 surface expression and the resulting retention of T cells in peripheral tissue. Together these data identify CD103+CD8+ T cells in breast tumors and NCBTs as TRMs and highlights their restricted localization to peripheral tissue sites.
CD8+ TIL CD103 and CD69 expression patterns differ by tumor localization. Flow cytometry data of fresh tumor tissues revealed that CD8+ TILs comprised 3 main populations of T cells: CD69+CD103+, CD69+CD103–, and CD69–CD103–. CD69 has been shown induce T cell retention in peripheral tissues (14), but the localization of T cells within the tumor microenvironment has not been explored, we believe, within the context of CD69 and CD103 expression. FFPE tumor samples were assessed by QIF for CD69, CD103, and CD8 costaining (Figure 3 and representative images in Supplemental Figure 5). As expected, CD69+CD103+ cells were highly enriched in cancer islands relative to stroma. CD69+CD103– cells, however, were somewhat more evenly split between stroma and cancer islands although trending toward cancer island enrichment. Intriguingly, CD69–CD103–CD8+ T cells were found almost exclusively in the stroma. In agreement with our flow cytometry data, QIF rarely identified CD69–CD103+CD8+ T cells, supporting the use of CD103+CD8+ phenotyping as a marker of CD8+ TRMs for larger patient cohort analysis.
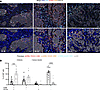 Figure 3
Figure 3CD103 and CD69 expression by CD8+ TILs varies by localization within the tumor microenvironment. FFPE tumor tissues were assayed by QIF for expression of both CD69 (shown in orange) and CD103 (shown in red) in the context of CD8 (shown in cyan) staining within the stroma and cancer islands (shown in gray, CK staining). (A) Representative merged composite images and identified T cell phenotypes (red dots, CD8+CD69+CD103+; orange dots, CD8+CD69+CD103–; cyan dots, CD8+CD69–CD103–; gray dots, CK+) are shown. Scale bars: 50 μm. Single-channel representative images can be found in Supplemental Figure 5. (B) The percentage of CD8+ T cells within stroma and cancer islands expressing either CD69 or CD103 was quantified as shown. Tumor samples n = 13. Significance was calculated using 1-way ANOVA and Holm-Šídák multiple-comparisons tests. ***P < 0.001; ****P < 0.0001.
CD8+ TRMs have a similar functional capacity compared to non-TRM tissue CD8+ T cells. Next we investigated the functional status of CD103+CD8+ TRMs and CD103–CD8+ non-TRMs from breast tumors and NCBTs. CD103+CD8+ TRMs have previously been shown to express elevated levels of checkpoint molecules, such as programmed cell death protein 1 (PD-1) (23, 26). We examined PD-1 expression in the context of CD103 and CD69 expression on CD8+ T cells in single-cell suspensions of fresh tumor and NCBT samples by flow cytometry (Supplemental Figure 6). Higher frequencies of both CD69+CD103+ and CD69+CD103– cells expressed PD-1 compared with CD69–CD103– cells in tumor tissue and NCBT. Thus, PD-1 expression was more associated with CD69 expression by CD8+ T cells rather than specifically CD8+ TRMs.
Next we interrogated cytokine production capacity of CD8+ TIL subsets for expression of IFN-γ, TNF-α, and IL-2 by intracellular flow cytometry (Figure 4, A and B). Memory CD45RA–CD8+ T cells were gated on CD69+CD103+ TRMs, CD69+CD103– cells, and CD69–CD103– cells to compare cytokine production between these subsets. CD103+CD8+ T cells in both breast tumors and NCBTs exhibited similar cytokine production patterns as compared to CD69+CD103– and CD69–CD103–CD8+ T cells in the same tissues in response to both PMA/ionomycin stimulation (Figure 4C) and anti-CD3 stimulation (Supplemental Figure 7). Furthermore, similar frequencies of CD8+ T cell subsets from both ER+ and TNBC tumors produced IFN-γ, TNF-α, and IL-2 (Supplemental Figure 4C). To quantitatively evaluate an overall functional capacity for each CD8+ T cell subset in both tissue types, we calculated a polyfunctionality index, which accounts for the ability of a T cell population to produce 1, 2, or 3 cytokines. No significant differences in polyfunctional capacity between CD8+ T cell subsets within breast tumors or NCBTs were observed (Figure 4D). Thus, the specialized function of CD8+ TRMs in human breast tumors is not attributed to differential or enhanced cytokine production potential.
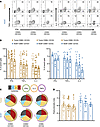 Figure 4
Figure 4CD8+ TRMs have similar cytokine production capacity as other tissue-infiltrating CD8+ T cells. CD8+ T cells from tumors and NCBTs were assayed by intracellular cytokine staining for IFN-γ, IL-2, and TNF-α following stimulation with PMA and ionomycin. (A) Cytokine production was assayed within CD69+CD103+, CD69+CD103–, and CD69–CD103– cells as shown. (B) Cytokine production profiles for different T cell populations are shown as percentage of cells within each gate producing a given cytokine. (C) Cytokine production is also summarized as the number (pie slice) and type (pie arcs) of cytokines produced by each population as Simplified Presentation of Incredibly Complex Evaluations (SPICE) pie charts. (D) A calculated polyfunctionality index representing the capacity of a given population to produce multiple cytokines is shown. Each symbol represents data from a unique patient sample. For individual cytokine data presented tumor n = 19; NCBT n = 9. For polyfunctional index data tumor n = 18; NCBT n = 9. Significance was calculated using 1-way ANOVA and Holm-Šídák multiple-comparisons tests. No differences (P > 0.05) in cytokine production capacity were identified between any groups.
CD103+CD8+ T cell infiltration of cancer islands in human breast tumors associates with relapse-free outcome. Recurrence in patients with TNBC peaks 3 years after surgery and rapidly declines thereafter (27). We therefore divided our TNBC FFPE cohort samples into relapse and relapse-free groups. Relapse patients were defined as having recurrence within 3 years of primary tumor removal, and relapse-free patients were defined as having no recurrence for at least 5 years. QIF results of CD8 and CD103 costaining were then assessed in the context of these outcomes (Figure 5 and representative images in Supplemental Figures 8 and 9).
 Figure 5
Figure 5CD103+CD8+ TRM infiltration in breast tumors associates with relapse-free outcome in patients. FFPE tumors assayed for the presence of CD103+CD8+ resident memory T cells by QIF were grouped according to relapse and relapse-free status. (A) Representative phenotype-mapped images with CK (gray), DAPI (blue), and T cell type phenotypes (cyan dots, CD103–CD8+; red dots, CD103+CD8+) are shown. Scale bars: 50 μm. Summarized data of CD8+ T cell densities (B) or CD103+CD8+ and CD103–CD8+ T cell densities (C) within total tissue, cancer islands, and stroma are shown. Each symbol represents data from a unique patient sample. Tumor samples n = 25. Significance was calculated using Holm-Šídák multiple-comparisons tests. ns, P > 0.05; **P < 0.01; ***P < 0.001.
As expected, a positive trend for association between increased overall density of CD8+ T cells in tumor tissue and RFS (P = 0.108) was identified (Figure 5B). However, increased density of CD8+ T cells localized specifically within cancer islands associated significantly with RFS (P = 0.002). This strong association with RFS was specific to the density of CD8+ T cells in cancer islands because the density of CD8+ T cells in stromal areas was less significantly associated with RFS (P = 0.158).
We next examined whether localization of specifically CD103+CD8+ TRMs to cancer islands was associated with RFS in our TNBC patient cohort (Figure 5C). The density of CD103+CD8+ TRMs in tumors from relapse-free patients was significantly higher than in tumors from relapse patients. Importantly, the density of TIL CD8+CD103– T cells did not associate with RFS and showed no significant differences in either the cancer islands or stroma of tumors. Higher densities of CD8+CD103+ TRMs were identified in relapse-free patients both in the cancer islands and in the stroma of tumors, suggesting that CD8+ TRMs are enriched in but not necessarily restricted to cancer islands, allowing for migration and surveillance by CD8+ TRMs within the tumor microenvironment.
The measurement of CD8+ T cell density within cancer island tissue regions does not account for CD8+ T cells localized in the stroma that are very near cancer islands. To address this, we conducted spatial analysis of CD8+ T cells with nearest neighbor analysis in respect to CK+ cancer cells (Figure 6). This allowed us to examine the spatial relationship between epithelial cells and all CD8+ T cells within close proximity (0–25 μm), medium proximity (25–50 μm or 50–75 μm), and far (>100 μm) from CK+ cancer cells. As expected, in all tumors, the frequency of CD8+CD103+ T cells within 0–25 μm of cancer cells was significantly higher than the frequency of CD8+CD103– T cells within 0–25 μm of cancer cells (Figure 6B). In contrast, CD8+ T cells far (>100 μm) from cancer cells were more enriched with CD8+CD103– T cells. Finally, a higher frequency of CD8+CD103+ T cells localized within 0–25 μm of cancer cells was significantly associated with RFS in the patients with TNBC (Figure 6C). Strikingly, there was no significant difference in the frequency of CD8+CD103– T cells localized within 0–25 μm of cancer cells between the relapse-free and relapse groups. Instead, the relapse group had a significantly higher fraction of CD8+CD103+ cells localized more than 100 μm from cancer cells, demonstrating that a lack of robust cancer island infiltration by CD8+ TRMs associates with poor prognosis in patients with TNBC.
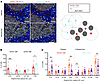 Figure 6
Figure 6Proximity of CD103+CD8+ TRMs and not CD103−CD8+ T cells to cancer cells in breast tumors associates with relapse-free outcome. FFPE tissues of relapse or relapse-free patients were assessed for the spatial relationship between CD103+CD8+ or CD103–CD8+ and CK cancer cells. (A) Representative phenotype-mapped images with CK+ (gray), DAPI (blue), and T cell type phenotypes (cyan dots, CD103–CD8+; red dots, CD103+CD8+) are shown with white lines representing nearest neighbor analysis of T cells within 25 μm of a CK+ cancer cell as depicted in a cartoon graphic. Spatial localization of CD103+CD8+ and CD103–CD8+ T cells within 0–25 μm, 25–50 μm, 50–100 μm, or more than 100 μm of the nearest CK+ cancer cell was assessed in all tumors (B) and is shown with regard to relapse and nonrelapse groups as scatter plots (C). Each symbol represents data from a unique patient sample. Tumor samples n = 25. Significance was calculated using Holm-Šídák multiple-comparisons tests. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Among the variables observed for total CD8+ T cells, CD8+CD103+ TRMs, and CD8+CD103– T cells in TNBC tumors, logistic regression analysis identified the density of cancer island CD8+CD103+ TRMs as the variable with the largest influence on predicting relapse-free outcome (Figure 7A). Addition of other variables in a multivariate model made no further significant contribution. The density of CD8+ T cells in stroma correlated weakly with the density of CD8+ T cells in cancer islands (rho = 0.34; P < 0.1) (Figure 7B). Thus, tumor stroma infiltration of CD8+ T cells, as described in noninflamed tumors, does not necessarily result in cancer island infiltration of CD8+ T cells. However, expression of CD103 by CD8+ TRM TILs results in significant cancer island infiltration and retention, and most important, improved prognostic outcome. The density of CD8+ T cells in cancer islands and the density of CD8+CD103+ TRMs in cancer islands were highly correlated (rho = 0.97; P < 0.0001), while the density of total tumor CD8+ T cells and the density of cancer island CD8+CD103+ TRMs were less correlated (rho = 0.53; P < 0.01). Together, these data identify TRM CD8+ infiltration of cancer islands as both a strong predictor of relapse-free outcome in patients with breast cancer and as a key aspect of immune-inflamed tumors.
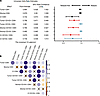 Figure 7
Figure 7CD8+CD103+ TRMs are the strongest indicator of RFS. (A) Univariate odds ratio assessment per increment of 10 cells/mm2 is shown, with respect to CD8+ T cells (black lines), CD103+CD8+ T cells (red lines), and CD103–CD8+ T cells (blue lines) in total tumor, stroma, or cancer islands. (B) A correlation matrix of all variables is also presented in which rho is displayed on a color scale and nonsignificant correlations (P > 0.05) are marked out with an X.
-
Discussion
In this study, we show that the density of CD8+ T cells within cancer islands is more significantly associated with relapse-free outcome than CD8+ T cells in overall tumor tissue or tumor stroma. Cancer island–localized CD8+ TILs are composed of CD103+ TRMs, which make up nearly half the total CD8+ T cell population within breast tumors and are the majority of CD8+ T cells in NCBT. Although CD103+CD8+ TRMs do not demonstrate unique functional capacity as measured by cytokine production, they do demonstrate enhanced ability to spatially localize near and among epithelial cells, including cancer cells. This localization of CD103+CD8+ TRMs to cancer islands in TNBC tumors is significantly associated with RFS (P = 0.0037; Figure 5) and furthers our current understanding of the association between CD8+ TILs and RFS in breast cancer.
Pathological assessment of stromal localized TILs, as opposed to cancer island–localized TILs, is currently one of the primary immune evaluation parameters in breast tumors (28). Restricting analysis to tumor stromal areas rather than cancer islands is due to generally less difficulty in assessment by pathologist review of H&E slides. A likely contributor to this is the significantly higher density of CD8+ infiltration in stromal areas as compared with cancer islands (Figure 1). However, density of CD8+ TILs within cancer islands proved to be highly associated with RFS even in our relatively small cohort, while density of CD8+ TILs within stroma was less so (Figure 5). Strikingly stroma infiltration of CD8+ TILs had no correlation with cancer island infiltration of CD8+ TILs. Additionally, our observation that stromal CD8+ TILs are primarily CD69–CD103–, which we show to have the lowest frequency of PD-1 expression, leads us to question the role of stroma CD8+ TILs in antitumor immunity. Clearly there is a need for a further understanding of the dynamic infiltration of tumor tissues by CD8+ T cells in the context of phenotypic changes.
Our data suggest that assessment of cancer island infiltration by CD8+CD103+ T cells may be useful in identifying CD8+ TRMs, predicting outcome, and assessing tumor T cell infiltration. CD103 expression in breast tumor tissues has been correlated with RFS (29). However, CD103 staining may also be attributed to expression on dendritic cells, regulatory T cells, and other lymphocytes (30–32), highlighting the need for multiplex staining for studies such as these. Quantitative immunostaining approaches offer the advantage of accurate cell subset identification, enumeration, and spatial localization within tumor microenvironments that is superior to analysis of fresh tissue digests (33). Indeed, spatial localization of CD8+ TRMs to cancer islands and their densities within tumors would not necessarily be readily identifiable by other technologies, such as flow cytometry or RNA-sequencing approaches.
A TRM gene expression signature based on single-cell sequencing has recently been found to associate with good prognosis in TNBC public data sets (22). Complementary to our QIF data, Savas et al. showed that increased expression of a CD8+ TRM transcriptional signature was of greater prognostic value than a CD8+ signature alone in TNBC. Our data further demonstrate that increased density of CD8+ TRMs in both the cancer islands and stroma of tumor tissues makes up an immune-inflamed tumor profile. Current standard of care for primary TNBC tumors includes neoadjuvant chemotherapy before surgical resection, leading to limited availability of fresh untreated TNBC tumor samples for research, as is evident by the predominance of ER+ tumor samples in our fresh tissue studies. TNBC was chosen to study the role of CD8+ TRMs in patient prognosis because of the known positive association between CD8+ TILs and relapse-free outcome (4). Whether our observation of the relationship between cancer island infiltration by CD8+ TRMs and relapse-free outcome in our TNBC cohort is related to overall higher CD3+ and CD8+ TILs observed in TNBC relative to other breast cancer cohorts is unclear (34). A larger cohort, both validating our results and extending our findings regarding CD8+CD103+ TRMs to other subsets of invasive breast cancer, especially hormone receptor–positive disease, is needed.
Recent single-cell sequencing efforts have shown breast tumor CD8+ TILs to be a heterogeneous population of T cells in various states of activation and differentiation (35). Our data demonstrate that CD8+ TRMs are a major T cell population within human breast tumors. TRMs are a subset of T cells, either CD8+ or CD4+, that are retained and long-lived in peripheral tissues (36–38). Originally identified in murine models of pathogen infection, TRMs enable critical and rapid protective immunity at peripheral tissue sites (39). TRMs facilitate this upon reencountering antigens by secreting cytokines, such as IFN-γ and TNF-α, that elicit further T cell recruitment, mature dendritic cells, activate natural killer cells, and initiate tissue-wide inflammatory pathways (40, 41). The developmental pathway of tumor TRMs and their mechanistic relationship with relapse-free outcome is not yet clear. Because tumor tissues are removed by surgical excision, the prognostic benefit of CD8+ TRMs likely reflects a greater systemic immunity involving circulating CD8+ T cells with shared antigen specificity or at least tumor specificity with tumor CD8+ TRMs. Although tumor specificity of breast tumor TILs and TRMs has not been formally proved, T cell receptor repertoire analysis of matched patient samples has shown that certain clonotypes are enriched in the tumor and not in NCBT (42, 43). TRMs may also develop in peripheral tissues independent of antigen presence because of the presence of various cytokines common in tumor microenvironments, such as TGF-β, TNF-α, IL-33, and IL-15 (44, 45). Recent work has elegantly shown that TIL TRMs can be composed of both tumor-specific and tumor-nonspecific, “bystander” cells (46). The presence of bystander TRMs specific for viral antigens in tumor tissues highlights the role of inflammatory cues in promoting TRM formation and cautions against assuming TRMs in tumor tissues are necessarily tumor specific. Further studies to identify antigen specificity of breast tumor TILs will allow for a greater understanding of tumor immune composition and the role of T cells in patients with breast cancer.
Vaccine-induced development of CD8+ TRMs demonstrated effective protection against tumor induction in a murine tumor model (47). Additionally, the induction or presence of CD103+CD8+ TRMs has been shown to enhance response to checkpoint blockade therapy in both preclinical and clinical research settings (26, 48). Although current response rates to checkpoint blockade therapies have been relatively low in patients with breast cancer, focusing on patients with a significant density of intraepithelial CD8+ TRMs may enable more significant responses to immunotherapeutic interventions (49). Thus, a greater understanding of TRM formation, function, and therapeutic benefit in breast tissues may enable immunotherapy strategies for patients with breast cancer. Finally, evaluating the presence of CD103+CD8+ TRMs within cancer islands may prove useful for determining patient prognosis and efficacy of various immune-modulating therapies in patients with breast cancer.
-
Methods
Human samples. Fresh surgically excised tumors and NCBTs were obtained from consented patients with breast cancer undergoing standard-of-care treatment at City of Hope. Patient tissue characteristics are summarized in Supplemental Table 2. Scoring of tumor tissue receptor expression was performed by clinical pathologists. NCBTs were composed of tissue from prophylactic mastectomies, the contralateral breast from patients with breast cancer, or tumor-adjacent tissue, as summarized in Supplemental Table 3. Because of limited cell numbers obtained from patient tumor samples, not all analyses shown were performed on all samples. Tissue samples were provided by the City of Hope Biospecimen Repository, which is funded in part by the National Cancer Institute. Other investigators may have received specimens from the same patients.
For evaluation of the relationship between outcome and the presence of CD103+CD8+ TRMs in breast cancer patient tumors, we compiled FFPE tissues from 25 patients with TNBC who had been followed for at least 5 years. Clinical characteristics are summarized in Supplemental Table 1. All patients were untreated before surgical removal of tumor tissue, and all patients were treated with similar (doxorubicin, cyclophosphamide, paclitaxel) chemotherapy regimens following surgery. For analysis of CD103+CD8+ TRMs in NCBTs, FFPE tissues were obtained from age-matched healthy donors with no history of breast cancer. Tissue samples were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. Other investigators may have received specimens from the same subjects.
Sample processing. Patient peripheral blood was obtained by venipuncture using heparin collection tubes, transported at room temperature from the clinic to the lab, and processed within 6 hours of drawing. PBMCs were isolated via Ficoll-Paque separation (GE Healthcare) following the manufacturer’s instructions. Breast tumor and noncancerous tissue specimens were collected by surgical resection and collected in tubes containing cold RPMI medium (Life Technologies, Thermo Fisher Scientific) and transported on ice to the laboratory for processing within 1 hour of surgery. Tissues were minced into pieces, mechanically dissociated with a gentleMACS Dissociator (Miltenyi Biotec), and enzymatically treated with 0.2 Wunsch U/mL Liberase (Roche) and 10 units/mL DNAse (MilliporeSigma) in RPMI medium for up to 1 hour as needed. If necessary, red blood cell (RBC) lysis was performed using RBC Lysis Buffer (BioLegend).
Flow cytometry. Single-cell suspensions were stained at room temperature in 2% FBS in PBS. For cytokine production assays, cells were stimulated with 50 ng/mL PMA (MilliporeSigma) and 1 μg/mL ionomycin (MilliporeSigma) or 1 μg/mL of anti-CD3 (clone OKT3, BioLegend) in the presence of GolgiPlug (BioLegend) for 4 hours. Overnight fixation as needed was performed with IC Fixation Buffer (eBioscience). Fixation and permeabilization were performed with BD Cytofix/Cytoperm buffers for intracellular cytokine staining. Antibody cocktails were diluted in Brilliant Violet Buffer (BD Biosciences) when necessary. Samples were acquired using a BD Fortessa using FACSDiva 6.1.3. Photomultiplier tube voltages were set using BD CS&T beads. Compensation was calculated using single stained OneComp compensation beads (eBioscience). Samples were stained with fluorescently tagged antibodies detailed in Supplemental Table 4. Antibodies were titrated for optimal signal to noise ratio before use. Flow cytometry analysis was performed using FlowJo vX. All samples were gated on single cells, lymphocytes, and CD3+CD8+ populations. Contour plots shown display 5% probability. The polyfunctionality index equation was applied as described by Larsen et al. (50). The polyfunctionality index was implemented in R (version 3.3.2) to take SPICE-formatted csv files as inputs and output a txt file with the polyfunctionality index of each sample.
Real-time PCR. CD8+ T cells were isolated from tumor tissue via culture of tissue fragments with high-dose IL-2 as previously described (51). Cells were collected and sorted into CD69+CD103+ and CD69+CD103– populations using an ARIA III flow cytometry sorting instrument (BD Biosciences). To serve as non-TRM controls, circulating memory cells were extracted from PBMCs of age- and sex-matched healthy donors using an EasySep Human Memory CD8+ T cell Enrichment Kit (STEMCELL Technologies). RNA from isolated cells was extracted with the RNeasy Micro Kit (Qiagen), from which cDNA was synthesized using the SuperScript VILO kit (Invitrogen). Quantitative real-time PCR reactions were run using Power SYBR Green PCR Master Mix (Applied Biosystems) for 40 cycles. RNA expression in CD69+CD103+ and CD69+CD103–CD8+ T cells was normalized relative to expression of ACTB and evaluated compared to gene expression in circulating CD8+ memory T cells from 3 healthy donors. Primers are described in Supplemental Table 4.
Immunofluorescent staining. FFPE specimens were cut into 3- to 5-μm sections and baked on glass slides. The FFPE slides were deparaffinized in xylene and then rehydrated in decreasing gradations of ethanol. Heat-induced epitope/antigen retrieval was performed in EnVision FLEX Target Retrieval Solution, High pH (pH 9) (K8004/5, Agilent) or AR6 buffer (pH 6) (PerkinElmer) using a microwave oven (Sharp Carousel). Blocking was performed for 10 minutes using Antibody Diluent, Background Reducing (S3022, Agilent), to minimize nonspecific background staining. Primary antibodies, as shown in Supplemental Table 4, were incubated for 1 hour on a shaker at room temperature, detected by a horseradish peroxidase–conjugated (HRP-conjugated) secondary antibody (Mach 2 Rabbit or Mouse HRP-Polymer), and then immunofluorescently labeled using the Opal 7-color fluorescence kit (PerkinElmer). Heat-mediated antigen retrieval using a microwave was performed in between serial stains of antibodies of interest. Cell nuclei were stained with DAPI (PerkinElmer), and the slides were mounted with ProLong Gold Antifade Reagent (P36930, Thermo Fisher Scientific).
Multispectral imaging and QIF image analysis. Tissue sections were whole-slide scanned using the Vectra 3.0 System (PerkinElmer) to capture fluorescent spectra of original magnification ×10 and ×20 images in 5 channels (DAPI, FITC, Cy3, Texas red, Cy5). Using Phenochart whole-slide reviewer (PerkinElmer), regions of interest (ROIs) were selected, and 25% of the images within the ROIs were systemically gridded and selected to unbiasedly capture tissue heterogeneity for further analysis. Images of single stained tissues and unstained tissues were used to extract the fluorescent spectrum of each fluorophore and tissue autofluorescence in the ×20 images to create a spectral library to perform multispectral unmixing using inForm Cell Analysis (PerkinElmer). inForm Cell Analysis or QuPath image analysis software was used for cell segmentation, tissue segmentation, and cell phenotyping (52). Supplemental Figure 3 summarizes the multispectral imaging and quantitative image analysis approach summarized above.
Cell phenotype spatial analysis. Spatial data analysis was performed by inputting cell phenotype x and y coordinates into a K-nearest neighbor algorithm to determine nearest neighbor cells of a particular phenotype within 50 pixels (25 μm) of any cell. Images were recoordinated so that all the cells in the tissue could be represented on the same set of coordinate axes. The nearest neighbor pairs of interest were CD103+CD8+ cell to CK+ cell and CD103–CD8+ cell to CK+ cell. The TRM and non-TRM T cells within 25 μm of a cancer cell were counted and then normalized by tumor area in square millimeters. Spatial analysis, spatial analysis–related figures, and spatial analysis calculations were performed in R version 3.4.3. R scripts were written in house and are available at https://github.com/TravisYTu/TRM_NN The K-nearest neighbor algorithm was imported from the RANN package in R.
Statistics. Analysis and presentation of distributions were performed using SPICE version 5.1, downloaded from https://niaid.github.io/spice/ (53). Graphs and statistical analyses were generated using GraphPad Prism 7.02. Statistics described were generated using 2-tailed Student’s t tests, 1-way ANOVAs, and Holm-Šídák multiple-comparisons tests. Correlation matrices were performed using the R package corrplot. Logistic regression and odds ratio analysis were performed using SAS software. Calculated P values are displayed as *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001. P values less than 0.05 were considered significant. For all graphs, the mean is represented by a bar and the error bars represent SEM.
Study approval. Fresh tumor and peripheral blood were obtained from patients who gave institutional review board–approved (IRB-approved) written informed consent before inclusion in the study (City of Hope IRB 05091, IRB 07047, and IRB 14346).
-
Author contributions
CAE and PPL designed research studies. CAE, CA, TYT, AR, RW, GS, YH, MHL, DLS, SS, MSN, TFH, and WG conducted experiments, acquired data, and analyzed data. JHY, LK, JM, SY, and YY provided clinical sample support. PF and CR assisted with statistical analysis. CAE and PPL wrote the manuscript.
-
Acknowledgments
The authors would like to thank Michele Kirschenbaum (City of Hope) for obtaining patient consent, procuring tissue samples, and obtaining clinical information and Eliza Barragan (City of Hope) for assistance with patient clinical information. We thank Sabina Muend (City of Hope) and Robert Zhang (City of Hope) for critical reading of this manuscript. We especially thank our patient tissue donors and our breast cancer patient advocate Susie Brain (advocate for Department of Defense Breast Cancer Research Program). This work was supported by the Department of Defense Breast Cancer Research Program award (W81XWH-11-1-0548), a Stand Up to Cancer–Breast Cancer Research Foundation Team award (2015-001), and V Foundation. Research reported in this publication included work performed in the Analytical Cytometry Core and the Pathology Research Services Core supported by the National Cancer Institute of the NIH under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Address correspondence to: Peter P. Lee, City of Hope Comprehensive Cancer Center, Beckman Center, Room 5117, 1500 East Duarte Road, Duarte, California 91010, USA. Phone: 626.218.2519; Email: plee@coh.org.
-
Footnotes
Conflict of interest: YY has contracted clinical trials and research projects sponsored by Merck, Eisai, Novartis, Genentech, and Pfizer.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(19):e130000.https://doi.org/10.1172/jci.insight.130000.
-
References
- Denkert C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28(1):105–113.View this article via: PubMed Google Scholar
- Aaltomaa S, et al. Lymphocyte infiltrates as a prognostic variable in female breast cancer. Eur J Cancer. 1992;28A(4–5):859–864.View this article via: PubMed Google Scholar
- Adams S, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32(27):2959–2966.
- Ali HR, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543.
- Ruffell B, Au A, Rugo HS, Esserman LJ, Hwang ES, Coussens LM. Leukocyte composition of human breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2796–2801.
- Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(Suppl 2):S60–S64.View this article via: PubMed Google Scholar
- Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752.
- Carey LA, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334.
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330.
- Müller P, et al. Trastuzumab emtansine (T-DM1) renders HER2+ breast cancer highly susceptible to CTLA-4/PD-1 blockade. Sci Transl Med. 2015;7(315):315ra188.
- Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571.
- Strauch UG, et al. Integrin αE(CD103)β7 mediates adhesion to intestinal microvascular endothelial cell lines via an E-cadherin-independent interaction. J Immunol. 2001;166(5):3506–3514.
- Bankovich AJ, Shiow LR, Cyster JG. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J Biol Chem. 2010;285(29):22328–22337.
- Mackay LK, et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol. 2015;194(5):2059–2063.
- Masopust D, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207(3):553–564.
- Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483(7388):227–231.
- Laidlaw BJ, et al. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 2014;41(4):633–645.
- Hofmann M, Pircher H. E-cadherin promotes accumulation of a unique memory CD8 T-cell population in murine salivary glands. Proc Natl Acad Sci U S A. 2011;108(40):16741–16746.
- Webb JR, Milne K, Watson P, Deleeuw RJ, Nelson BH. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin Cancer Res. 2014;20(2):434–444.
- Djenidi F, et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol. 2015;194(7):3475–3486.
- Malik BT, et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci Immunol. 2017;2(10):eaam6346.
- Savas P, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24(7):986–993.
- Webb JR, Milne K, Nelson BH. PD-1 and CD103 are widely coexpressed on prognostically favorable intraepithelial CD8 T cells in human ovarian cancer. Cancer Immunol Res. 2015;3(8):926–935.
- Dumauthioz N, Labiano S, Romero P. Tumor resident memory T cells: new players in immune surveillance and therapy. Front Immunol. 2018;9:2076. View this article via: PubMed Google Scholar
- Mackay LK, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;14(12):1294–1301.
- Wang P, et al. CD103(+)CD8(+) T lymphocytes in non-small cell lung cancer are phenotypically and functionally primed to respond to PD-1 blockade. Cell Immunol. 2018;325:48–55.
- Dent R, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–4434.View this article via: PubMed Google Scholar
- Salgado R, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–271.
- Wang ZQ, Milne K, Derocher H, Webb JR, Nelson BH, Watson PH. CD103 and intratumoral immune response in breast cancer. Clin Cancer Res. 2016;22(24):6290–6297.
- Annacker O, et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J Exp Med. 2005;202(8):1051–1061.
- Allakhverdi Z, et al. Expression of CD103 identifies human regulatory T-cell subsets. J Allergy Clin Immunol. 2006;118(6):1342–1349.
- Edelson BT, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J Exp Med. 2010;207(4):823–836.
- Steinert EM, et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 2015;161(4):737–749.
- Miyan M, Schmidt-Mende J, Kiessling R, Poschke I, de Boniface J. Differential tumor infiltration by T-cells characterizes intrinsic molecular subtypes in breast cancer. J Transl Med. 2016;14(1):227.
- Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–1308.e36.
- Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291(5512):2413–2417.
- Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol. 2016;16(2):79–89.
- Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41(6):886–897.
- Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10(5):524–530.
- Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346(6205):98–101.
- Ariotti S, et al. T cell memory. Skin-resident memory CD8(+) T cells trigger a state of tissue-wide pathogen alert. Science. 2014;346(6205):101–105.
- Beausang JF, et al. T cell receptor sequencing of early-stage breast cancer tumors identifies altered clonal structure of the T cell repertoire. Proc Natl Acad Sci U S A. 2017;114(48):E10409–E10417.
- Munson DJ, et al. Identification of shared TCR sequences from T cells in human breast cancer using emulsion RT-PCR. Proc Natl Acad Sci U S A. 2016;113(29):8272–8277.
- Casey KA, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. 2012;188(10):4866–4875.
- Mackay LK, et al. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci U S A. 2012;109(18):7037–7042.
- Simoni Y, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557(7706):575–579.
- Nizard M, et al. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat Commun. 2017;8:15221. View this article via: PubMed Google Scholar
- Enamorado M, et al. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat Commun. 2017;8:16073. View this article via: PubMed Google Scholar
- Nanda R, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016;34(21):2460–2467.
- Larsen M, Sauce D, Arnaud L, Fastenackels S, Appay V, Gorochov G. Evaluating cellular polyfunctionality with a novel polyfunctionality index. PLoS One. 2012;7(7):e42403.
- Turcotte S, et al. Phenotype and function of T cells infiltrating visceral metastases from gastrointestinal cancers and melanoma: implications for adoptive cell transfer therapy. J Immunol. 2013;191(5):2217–2225.
- Bankhead P, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878.
- Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79(2):167–174.View this article via: PubMed Google Scholar
- Denkert C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28(1):105–113.
-
Version history
- Version 1 (August 29, 2019): In-Press Preview
- Version 2 (October 3, 2019): Electronic publication










