Research ArticleOncology Free access | 10.1172/jci.insight.87380
PML promotes metastasis of triple-negative breast cancer through transcriptional regulation of HIF1A target genes
Manfredi Ponente,1,2 Letizia Campanini,1,2 Roberto Cuttano,1 Andrea Piunti,1 Giacomo A. Delledonne,1 Nadia Coltella,1 Roberta Valsecchi,1 Alessandra Villa,3 Ugo Cavallaro,3 Linda Pattini,4 Claudio Doglioni,5 and Rosa Bernardi1
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Ponente, M. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Campanini, L. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Cuttano, R. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Piunti, A. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Delledonne, G. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Coltella, N. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Valsecchi, R. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Villa, A. in: PubMed | Google Scholar
1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by
Cavallaro, U.
in:
PubMed
|
Google Scholar
|

1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by
Pattini, L.
in:
PubMed
|
Google Scholar
|

1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by
Doglioni, C.
in:
PubMed
|
Google Scholar
|

1Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute,
2Vita-Salute San Raffaele University,
3Department of Experimental Oncology and Molecular Medicine Program, European Institute of Oncology, Milan, Italy.
4Department of Electronics, Information and Bioengineering, Politecnico di Milano, Italy.
5Department of Pathology, IRCCS San Raffaele Scientific Institute, Milan, Italy.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
Find articles by Bernardi, R. in: PubMed | Google Scholar
Published February 23, 2017 - More info
JCI Insight. 2017;2(4):e87380. https://doi.org/10.1172/jci.insight.87380.
Copyright © 2017, American Society for Clinical Investigation
Received: March 3, 2016; Accepted: January 10, 2017
-
Abstract
Elucidating the molecular basis of tumor metastasis is pivotal for eradicating cancer-related mortality. Triple-negative breast cancer (TNBC) encompasses a class of aggressive tumors characterized by high rates of recurrence and metastasis, as well as poor overall survival. Here, we find that the promyelocytic leukemia protein PML exerts a prometastatic function in TNBC that can be targeted by arsenic trioxide. We found that, in TNBC patients, constitutive HIF1A activity induces high expression of PML, along with a number of HIF1A target genes that promote metastasis at multiple levels. Intriguingly, PML controls the expression of these genes by binding to their regulatory regions along with HIF1A. This mechanism is specific to TNBC cells and does not occur in other subtypes of breast cancer where PML and prometastatic HIF1A target genes are underexpressed. As a consequence, PML promotes cell migration, invasion, and metastasis in TNBC cell and mouse models. Notably, pharmacological inhibition of PML with arsenic trioxide, a PML-degrading agent used to treat promyelocytic leukemia patients, delays tumor growth, impairs TNBC metastasis, and cooperates with chemotherapy by preventing metastatic dissemination. In conclusion, we report identification of a prometastatic pathway in TNBC and suggest clinical development toward the use of arsenic trioxide for TNBC patients.
-
Introduction
Metastasis is the leading cause of cancer-associated mortality. In breast cancer, it has been calculated that metastatic dissemination may begin early in the process of tumorigenesis, with disseminated micro-metastasis giving rise to life-threatening macro-metastases years or decades after initial diagnosis (1). In addition, tumor reseeding has been described from the primary tumor — as well as from established metastases — thus prompting the scientific community to devise innovative strategies to treat patients by targeting all aspects of metastatic dissemination: dormancy, colonization, and reseeding (2).
Triple-negative breast cancer (TNBC; representing 15%–20% of all breast cancers) is a tumor subtype that lacks expression of estrogen receptors (ER), progesterone receptors (PR), and HER2 receptors and is characterized by high rates of metastasis and poor overall survival (3). Because TNBC is a highly heterogeneous disease, targeted therapies are currently lacking and patients are treated with chemotherapy. Although their tumors are sensitive to chemotherapeutic regimens, TNBC patients have a high risk of developing disease relapse and resistance to treatment; therefore, new therapeutic strategies are urgently needed (3).
Interestingly, it was recently observed that, despite prominent genetic heterogeneity, TNBC displays deregulation of few transcriptional networks, which include activation of a hypoxia-dependent gene expression program (4–7). Hypoxia-inducible (HIF) transcription factors regulate cell adaptation to hypoxia and are often upregulated in tumors either by intratumoral hypoxia or through hypoxia-independent activation of specific oncogenic pathways (8). HIF factors regulate a variety of tumor-promoting mechanisms, including neo-angiogenesis, cancer stem cell maintenance, cell migration, and invasion (8). In breast cancer, high expression of HIF1A correlates with advanced disease and poor clinical outcome, and molecular studies have indicated that HIF1A promotes breast cancer metastasis by acting at multiple levels of the metastatic cascade (9, 10). More recently, normoxic expression of HIF1A and activation of hypoxia gene expression programs were reported specifically in TNBC (4–7), and it was suggested that targeting this pathway might provide a new therapeutic option for TNBC patients (4, 9).
The promyelocytic leukemia protein PML has been long described as a tumor suppressor that is downregulated in tumors and limits cancer progression by finely tuning a variety of tumor suppressive pathways (11). However, PML was recently found overexpressed in aggressive breast cancers, particularly of the triple-negative subtype, where it was suggested to function as an oncogene by promoting ATP production and cell survival, along with maintenance of breast cancer–initiating cells and tumor aggressiveness (12, 13).
In the present study, we show that PML is an HIF1A target gene and that high PML expression is promoted at least partly by HIF1A activation in TNBC. In TNBC patients, PML expression correlates with an HIF1A-dependent gene signature that contains a number of prometastatic genes acting at multiple levels within the metastatic cascade. Interestingly, we found that PML, in turn, regulates the expression of these genes and promotes TNBC metastatic features both in vitro and in vivo. As a consequence, targeting PML with arsenic trioxide, either alone or in combination with chemotherapy, effectively inhibits metastasis in TNBC. In sum, our results indicate that PML is a druggable target in TNBC and suggest that arsenic trioxide may be tested as a new antimetastatic agent in neo-adjuvant or adjuvant settings for a subset of breast cancer patients.
-
Results
PML is an HIF1A target gene in breast cancer. We have previously shown that, in prostate and kidney cancer, PML regulates HIF1A hypoxic accumulation and neo-angiogenesis (14, 15). While performing these studies, we had observed that PML levels increase in vivo in hypoxic conditions (14), suggesting that PML expression may be, in turn, regulated by HIF1A. In line with this hypothesis, in vitro experiments revealed that hypoxia mimetic compounds deferoxamine and cobalt chloride induced PML mRNA accumulation in an HIF1A-dependent manner in murine and human cells, albeit at later time points and to a lesser extent than the HIF1A target gene VEGF (Supplemental Figure 1, A and B, and data not shown; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.87380DS1). Analysis of the human PML promoter revealed 5 putative HIF-responsive elements (HREs) within 1 kb from the transcription start site, one of which (HRE#2) is conserved in the murine Pml promoter (Supplemental Figure 1C). Accordingly, mouse and human PML promoters drove luciferase expression following stable HIF1A expression or treatment with deferoxamine in an HIF1A-dependent manner (Supplemental Figure 1, D and E). Together, these data indicate that HIF1A regulates PML expression.
To investigate the relevance of PML transcriptional regulation by HIF1A, we focused on TNBC because PML was recently found overexpressed in this tumor subtype (12), where HIF1A is constitutively expressed through hypoxia-independent mechanisms (7, 16). To test the hypothesis that HIF1A promotes PML expression in TNBC, PML mRNA levels were first compared with a hypoxia-dependent gene expression signature that discriminates breast cancer patients with poor prognosis (17). PML was found significantly upregulated in breast cancer patients with a high hypoxia gene expression profile (Figure 1A) (17). In addition, PML mRNA levels were significantly higher in TNBC patients compared with normal tissue and other breast cancer subtypes (Figure 1B), consistently with the recently described upregulation of PML protein in TNBC (12). Finally, normoxic expression of HIF1A in TNBC cell lines (16) was accompanied by high PML mRNA and protein levels (Figure 1, C and D).
 Figure 1
Figure 1PML is regulated by HIF1A in TNBC. (A) PML mRNA expression (centered by mean and log10 transformed) in 295 early-stage breast cancer samples subcategorized for high and low expression of a hypoxia-responsive gene signature of 123 common hypoxia-dependent genes. (B) Mean PML mRNA expression in breast cancer samples characterized by TCGA Network and subcategorized as estrogen receptor positive (ER+, n = 401), HER2 positive (HER2+, n = 76), TNBC (n = 88), and normal tissue (n = 22). P values are: TNBC vs. normal, P = 0.0013; TNBC vs. HER2+, P = 3.08 × 10–6; TNBC vs. ER+ P = 1.61 × 10–14. (C) RT-PCR analysis of PML expression across different breast cell lines. Non-TNBC, and TNBC cells are indicated. Data represent mean values ± SEM of 2 independent experiments. Statistical analysis was performed by comparing mean PML expression in the 4 TNBC cell lines versus mean PML expression in the 3 non-TNBC cells, respectively. (D) Immunoblot of PML and HIF1A across breast cancer cell lines: estrogen and progesterone receptor positive (ER+/PR+) cells MCF7 and ZR-75, HER2+ overexpressing MDA-MB-361 cells, and TNBC cells SUM-149, BT-549, SUM-149, and MDA-MB-231. Cropped blots are surrounded by a black line and retain important bands. β-Actin is used as loading control. (E) RT-PCR analysis of the indicated genes in MDA-MB-231 and MCF7 cells stably transduced with a control shRNA (white dots) or an HIF1A shRNA (gray dots). Data represent mean values ± SEM of 3 independent experiments. (F) Fold enrichment over normalized IgG levels of DNA immunoprecipitated by control IgG (white dots) or anti-HIF1A antibody (gray dots) and amplified with primers spanning either the promoter region or the gene body of PML in MDA-MB-231 and MCF7 cells. Data represent mean values ± SEM of 3 independent experiments. **P < 0.01, ***P < 0.001. Student’s 2-tailed t test was used to determine statistical significance.
To test whether HIF1A directly regulates PML expression in TNBC, HIF1A was stably silenced in TNBC MDA-MB-231 cells and in ER+/PR+ MCF7 cells that express low levels of PML and HIF1A (Figure 1D). PML mRNA expression diminished along with expression of bona fide HIF1A target genes CA9 and GLUT1 only in MDA-MB-231 cells, while HIF1A silencing had no transcriptional effect on any of the genes analyzed in MCF7 cells (Figure 1E). Consistently, HIF1A specifically bound the PML promoter and not the gene body only in MDA-MB-231 cells (Figure 1F). To extend these observations, PML expression was analyzed in 4 additional breast cancer cells lines, representative of TNBC (SUM-159 and BT-549) and non-TNBC (MDA-MB-361 and ZR-75-30). HIF1A did not bind the PML promoter and did not regulate PML expression in non-TNBC cells (Supplemental Figure 2, B and C), while in the 2 additional TNBC cell lines, HIF1A silencing regulated PML levels only in SUM-159 cells and not in BT-549 (Supplemental Figure 2A), thus indicating that PML is also regulated through HIF1A-independent mechanisms in TNBC. In this regard, it was recently shown that STAT3 also controls PML expression in breast cancer (13), therefore suggesting that STAT3 and HIF1A may both lead to increased PML expression in TNBC.
Taken together, these data indicate that PML is an HIF1A target gene and that its expression is regulated, at least partly, by HIF1A in TNBC.
PML expression correlates with hypoxia-regulated metastasis genes in TNBC patients. To better characterize PML expression in relation to HIF1A activity within TNBC, a hypoxia-dependent gene signature correlating with poor prognosis in breast cancer (17) was analyzed in a larger cohort of breast cancer transcriptional profiles annotated by The Cancer Genome Atlas (TCGA) (4). Unsupervised hierarchical clustering segregated a group of patients (Figure 2A, black bar) where TNBC patients were strongly over-represented (P < 2.04 × 10–49), thus indicating that the expression pattern of hypoxia-dependent genes spontaneously discriminated TNBC samples. As expected, PML expression was significantly higher in this group of patients (Figure 2A: lower bar, P < 9.16 × 10–14). Interestingly, TNBC patients did not display upregulation of the entire hypoxia signature, but rather of a subset of hypoxia-regulated genes (Supplemental Table 1), thus suggesting that HIF1A may preferentially regulate selected target genes in TNBC cells. As expected, the TNBC hypoxia subsignature contained a number of genes that had been previously implicated in regulating metastasis in breast cancer, such as members of the PLOD and LOX family, PDK1, CA9, SOX4, CXCR4, ADM, ANGPTL4, IGFBP3, POU5F1, GPI, PGK1, SLC16A3, WIPF1, and ARRDC3 (18–32).
 Figure 2
Figure 2PML expression correlates with prometastatic hypoxia-regulated genes in TNBC. (A) Unsupervised clustering of 123 hypoxia signature genes applied to gene expression profiles of 547 TCGA breast cancer samples. Samples belonging to the cluster indicated by a black bar at the bottom are highly enriched in TNBC patients. At the bottom, PML expression along patients’ distribution shows significantly higher expression in TNBC patients. (B) Scatter plots describing the correlation of WIPF1, PLOD1, LOX, and ZEB2 expression with PML expression (Pearson correlation coefficient r and P values are indicated) in the 547 TCGA breast cancer samples. (C) Kaplan Meier analysis of relapse-free survival of 383 TNBC patients in correlation with PML mRNA expression.
In a parallel analysis, we found that 10 out of the top 15 HIF1A target genes whose expression most correlated with PML across breast cancer were included in the TNBC hypoxia subsignature (in bold in Supplemental Table 1). These comprised important regulators of breast cancer metastasis downstream HIF1A, such as PLOD1 and LOX, which promote remodeling of the extracellular matrix (ECM) and formation of premetastatic niches (18, 19), and WIPF1, which regulates actin cytoskeleton dynamics and cell migration and correlates with unfavorable prognosis in breast cancer (Figure 2B) (33, 34). In addition, the ZEB2 transcription factor, which induces epithelial to mesenchymal transition and breast cancer metastasis (35), was found within the 15 genes that most correlated with PML in breast cancer (Figure 2B) and was also upregulated in TNBC patients, albeit not significantly (FDR corrected P = 0.058).
Taken together, these results indicate that, in TNBC, PML is transcriptionally upregulated together with a hypoxia subsignature that includes several metastasis-promoting genes, and expression of some of these genes significantly correlates with PML expression in breast cancer. Consistently, PML expression inversely correlated with recurrence-free survival in TNBC patients (Figure 2C) (36).
PML controls the expression of metastasis genes in TNBC. Correlation of PML expression with HIF1A target genes driving metastasis (Figure 2B), along with its recently described protumorigenic function in TNBC (12), suggested that PML might be functionally involved in the regulation of these genes. To test this hypothesis, PML and HIF1A were constitutively silenced in breast cancer cell lines (Supplemental Figure 3, A and B). Downregulation of HIF1A or PML led to a general reduction in the expression of the prometastatic genes WIPF1, PLOD1, LOX, and ZEB2 in TNBC cell lines (Figure 3A and Supplemental Figure 3, A and B) but not in non-TNBC MCF7, MDA-MB-361, and ZR-75-30 cells (with the exception of PML regulating PLOD1 and LOX expression in ZR-75-30 cells; Figure 3B and Supplemental Figure 3, A and B). Of note, non-TNBC cells did not express detectable ZEB2 levels (not shown). These data indicate that the regulation of prometastatic HIF1A target genes by PML is tumor-subtype specific. Interestingly, PML downregulation in TNBC cells did not impair expression of HIF1A target genes involved in metabolic adaptation, like CA9 and GLUT1 (Supplemental Figure 3C), thus indicating that PML is involved in the regulation of a subset of HIF1A target genes in TNBC. Accordingly, CA9 and GLUT1 expression did not correlate with PML in breast cancer (not shown). Analogous results, although of smaller magnitude, were obtained in representative cells with a second shRNA (shPML#2; Supplemental Figure 3, D–F).
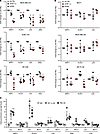 Figure 3
Figure 3PML regulates the expression of HIF1A target genes involved in metastasis. (A) RT-PCR analysis of the indicated genes in MDA-MB-231, SUM-159, and BT-549 TNBC cells stably transduced with a control shRNA (white dots), shRNA against HIF1A (gray dots), or shRNA against PML (red dots). Data represent mean values ± SEM of 3 independent experiments. (B) RT-PCR analysis of the indicated genes in MCF7, MDA-MB-361, and ZR-75-30 non-TNBC cells stably transduced with a control shRNA (white dots), shRNA against HIF1A (gray dots), or shRNA against PML (red dots). Data represent mean values ± SEM of 3 independent experiments. (C) Fold enrichment over normalized IgG levels of DNA immunoprecipitated by control IgG (white dots), HIF1A (light gray dots), and PML antibodies (dark gray dots) and amplified with primers spanning the regulatory regions of the indicated genes in MDA-MB-231 or MCF7 cells. Data represent mean values ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. Student’s 2-tailed t test was used to determine statistical significance.
Albeit not a transcription factor, PML has been reported to regulate transcription either by modulating the activity of transcription factors or by regulating chromatin organization (37, 38). Analysis of genome occupancy extrapolated from a large-scale ENCODE ChIP-seq experiment in human B lymphocytes (39) (www.encodeproject.org) revealed that PML binds the regulatory regions of 3 of the 4 metastasis genes here analyzed: WIPF1, PLOD1,and ZEB2 (Supplemental Figure 4A, blue boxes), while PML binding to LOX was not reported. Specific binding of PML to the regulatory regions of WIPF1, PLOD1, and ZEB2 identified by ENCODE and not to their gene bodies was confirmed in MDA-MB-231 cells (Figure 3C and Supplemental Figure 4, A and B). In addition, specific PML binding was revealed in the promoter region of the LOX gene, in proximity of the transcription start site (Figure 3C and Supplemental Figure 4, A and B). Consistently with lack of PML-mediated regulation (Figure 3B), PML did not bind the regulatory regions of these genes in MCF7 cells (Figure 3C). Notably, HIF1A occupied the same regulatory regions of the WIPF1, PLOD1, LOX, and ZEB2 genes in MDA-MB-231 cells, while binding was generally reduced in MCF7 cells (Figure 3C). Consistently, expression of these genes was globally lower, as was PML expression, in ER+ and HER2+ cell lines compared with TNBC cell lines in microarray data from the Cancer Cell Line Encyclopedia (http://software.broadinstitute.org/software/cprg/?q=node/11) (Supplemental Figure 5, A and B). Finally, in both MDA-MB-231 and MCF7 cells, PML did not bind the regulatory regions of the HIF1A target gene CA9 (Figure 3C), which was not regulated by PML silencing (Supplemental Figure 3C).
In summary, these results indicate that, in TNBC cells, PML participates to the regulation of HIF1A-dependent metastasis genes through a mechanism that may involve modulation of HIF1A transcriptional activity on chromatin.
PML promotes metastasis in TNBC cells. The functional consequences of PML expression in TNBC were next analyzed in vitro and in vivo. Overall, chronic PML silencing did not affect proliferation or survival of breast cancer cell lines (albeit some reduction in cell proliferation was observed in MDA-MB-231 cells; Supplemental Figure 6, A and B, and data not shown). In agreement with the specific regulation of metastasis genes, PML promoted features of metastasis only in TNBC cells in vitro. Cell migration was inhibited by PML silencing, specifically in TNBC cell lines, although the effect on SUM-159 cells was modest; this is perhaps due to their reduced migratory capacity in comparison with other TNBC cells (Figure 4A and Supplemental Figure 3G). In addition, cell invasion through matrigel, which only occurred by TNBC cells, was likewise inhibited by PML silencing (Figure 4B and Supplemental Figure 3H). Finally, in agreement with ZEB2 downregulation, PML silencing induced a phenotypic switch reminiscent of mesenchymal to epithelial reversion in TNBC cell lines, with decreased fibroblasts morphology and more evident cell-cell contacts (Figure 4C), although EMT markers like E-cadherin and N-cadherin did not change significantly (not shown).
 Figure 4
Figure 4PML regulates migration, invasion, and metastasis in TNBC cells. Wherever indicated, cells were stably transduced with a control shRNA (shCTRL, white dots) or PML-directed shRNAs (red dots). (A) Wound healing assays in the indicated cell lines. Data are expressed as wound area reduction compared with shCTRL cells. Data represent mean values ± SEM of 3 independent experiments. Pictures in the right panel are representative of MDA-MB-231 cells stably transduced with shCTRL (top) or PML shRNA (bottom) 24 hours after wound formation. (B) Invasion assays in the indicated cell lines. Data represent the number of cells per 20× field that invaded matrigel-coated transwells. Data represent mean values ± SEM of 3 independent experiments. (C) Bright-field images of the indicated cell lines. Scale bars: 200 μm. (D) Tumor volumes of MDA-MB-231 cells transduced with shCTRL or 2 shRNAs against PML (orange and red lines) and implanted in fat pads. Data represent mean values ± SEM; n = 5. Upper panels: PML IHC in representative shCTRL or shPML tumors. Scale bars: 50 μm. (E) Tumor weights from animals described in D. Data represent mean values ± SEM; n = 5. (F) Lung metastases in mice described in D. Left: number of metastatic foci per lung slide in the indicated animals. Data represent mean values ± SEM; n = 5. Right: H&E staining of lungs from representative shCTRL or shPML tumors. Black arrows indicate small metastatic foci in a representative shPML implanted mouse. Scale bars: 200 μm. In E and F, animals were sacrificed at 47 days (shCTRL) and 55 days (shPML) after cell implantation. *P < 0.05, **P < 0.01, ***P < 0.001.Student’s 2-tailed t test was used to determine statistical significance, except in panel F, where statistical analysis was performed using 1-way ANOVA (P = 0.0058) followed by Tukey’s post-hoc multiple comparison test.
To assess the regulation of metastasis by PML in vivo, MDA-MB-231 cells were implanted in the mammary fat pad of immunocompromized mice upon PML silencing. PML downregulation was maintained in vivo and delayed tumor growth (Figure 4D), in agreement with its reported regulation of cell survival and tumor-initiating capacity in breast cancer (12, 13). In addition, reducing PML expression led to a significant decrease in the number and size of metastatic foci to the lungs, even when primary tumors had reached similar weights of control tumors (Figure 4, E and F), thus demonstrating that, in TNBC, PML regulates metastasis independently of tumor growth retardation.
These results were confirmed in another model of spontaneous metastasis. Pml silencing in the mouse 4T1 cell line, which represents a mouse model of TNBC, led to decreased expression of the Wipf1, Plod1, Lox, and Zeb2 genes; decreased cell migration; and impaired metastasis in vivo (Supplemental Figure 7, A–D). Similar to the MDA-MB-231 experiment, Pml downregulation also impacted primary tumor growth in this model (Supplemental Figure 7C). However, metastasis formation was affected even if animals were sacrificed when their primary tumors reached similar sizes (Supplemental Figure 7, C and D).
In conclusion, these results indicate that PML is a regulator of metastasis in TNBC.
The PML targeting agent arsenic trioxide inhibits metastasis in TNBC. Arsenic trioxide is a pharmacological agent that induces PML degradation and is currently used to treat acute promyelocytic leukemia, where it promotes degradation of the PML-RARA fusion protein (40). Based on the function of PML as a prometastatic gene in TNBC, we tested the efficacy of arsenic trioxide as an antimetastatic agent for this cancer subtype. First, to test the antimetastatic function of arsenic trioxide, TNBC and ER+/PR+ cell lines were treated in vitro with concentrations of arsenic trioxide that suppressed PML expression without exerting overt cytotoxic effects for up to 48 hours (Supplemental Figure 8, A and B). Treatment with arsenic trioxide recapitulated PML silencing by specifically impairing the expression of WIPF1, PLOD1, LOX, and ZEB2 and inhibiting migratory and invasive features of TNBC cells MDA-MB-231 and SUM-159, while having no effect in MCF7 cells (Figure 5, A–C). Similar results were obtained with mouse 4T1 cells (Supplemental Figure 8, C–E).
 Figure 5
Figure 5Arsenic trioxide inhibits migration and invasion and cooperates with chemotherapy to block metastasis in TNBC. (A) RT-PCR analysis of the indicated genes in the indicated cell lines untreated (NT, white dots) or treated with the indicated doses of arsenic trioxide. Data represent mean values ± SEM of 3 independent experiments. (B) Wound healing assays in the indicated cell lines treated as in A. Data are expressed as wound area reduction compared with untreated cells and represent mean values ± SEM of 3 independent experiments. (C) Invasion assays of the indicated cell lines treated as in A. Data represent the number of cells per 20× field that invaded matrigel-coated transwells. Data represent mean values ± SEM of 3 independent experiments. (D) Tumor volumes of MDA-MB-231 cells implanted in fat pads and treated with PBS (NT, white bars) or with 8 mg/Kg arsenic trioxide. Treatment was started when tumors became palpable (black arrow). Four cycles of 5 treatments followed by 2 days off treatment were performed; n = 5. (E) Number of metastatic foci per lung slides in the indicated animals; n = 5. Data represent mean values ± SEM. (F) Tumor volumes of MDA-MB-231 cells implanted in fat pads and treated with PBS (NT, white bars), 4 mg/Kg arsenic trioxide, or 20 mg/Kg paclitaxel either alone or in combination according to the schedule described in the Methods section. Treatments were started when tumors became palpable (black arrow); n = 5. (G) Number of metastatic foci per lung slides in the indicated animals; n = 5. Data represent mean values ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Student’s 2-tailed t test was used to determine statistical significance except in panel G, where statistical analysis was performed using 1-way ANOVA (P < 0.0001) followed by Tukey’s post-hoc multiple comparison test.
In vivo, treatment with a high dose of arsenic trioxide delayed tumor progression and led to decreased expression of PML in tumors (Figure 5D and Supplemental Figure 8F). In addition, and similarly to PML silencing, arsenic trioxide reduced the metastatic burden from MDA-MB-231–derived tumors, even if arsenic-treated animals were sacrificed later than control animals to obtain primary tumors of similar weight (Figure 5E and Supplemental Figure 8F). In sum, these results indicate that arsenic trioxide mimics specific PML silencing in delaying tumor progression and impacting tumor metastasis in a model of TNBC.
Finally, to understand whether arsenic trioxide treatment would add therapeutic value to chemotherapy regimens used for TNBC patients, combination therapy was performed with arsenic trioxide and paclitaxel. Paclitaxel was administered with a protocol that blocks tumor progression and mimics conventional maximum dose regimens given to patients (41). Due to some in vivo toxicity of combined arsenic trioxide and paclitaxel treatment, arsenic trioxide dosage was lowered to 4 mg/Kg (Figure 5F). In these conditions, treatment with arsenic trioxide delayed growth of primary tumors less efficiently (Figure 5, D and F) but still significantly impaired metastatic spread (Figure 5G). Paclitaxel treatment alone arrested tumor growth (Figure 5F), as previously shown (41), and inhibited metastasis formation similarly to arsenic trioxide, although primary tumors from paclitaxel-treated animals were significantly smaller (Figure 5G and Supplemental Figure 8G), thus indicating that paclitaxel treatment does not specifically impairs metastatic spread. Significantly, when administered in combination with paclitaxel, arsenic trioxide treatment abated the metastatic potential of TNBC cells (Figure 5G). In summary, our results indicate that arsenic trioxide specifically inhibits metastatic spread in TNBC and may add therapeutic value to chemotherapeutic regimens that mainly act on primary tumors.
-
Discussion
Our study identifies the promyelocytic leukemia gene PML as a prometastatic gene in TNBC through transcriptional regulation of HIF1A target genes. We found that, in TNBC, PML expression is promoted at least partly by HIF1A, and PML in turn cooperates with HIF1A to support the expression of a number of metastasis-driving genes, thus stimulating cell migration and invasion. In vivo, PML silencing delays tumor growth and impairs metastatic dissemination in mouse models of TNBC, and arsenic trioxide — a PML targeting agent used to treat patients with promyelocytic leukemia — mimics PML suppression and acts as an antimetastatic agent in TNBC.
Although PML has long been described as a tumor suppressor, recent data indicates that it exerts oncogenic functions in specific contexts (11, 42). For example, PML is highly expressed in chronic myeloid leukemia (CML), where it promotes maintenance of leukemia stem cells (43, 44); therefore, PML downregulation via arsenic trioxide leads to CML eradication in combination with chemotherapy (43). More recently, PML was found upregulated in patients with aggressive breast cancer, particularly of the TNBC subtype, where it was shown to promote resistance to apoptosis through metabolic self-sufficiency (12). As our work was being evaluated for publication, a followup report of these findings revealed that PML also regulates tumor-initiating capacity in aggressive breast cancers by promoting SOX9 expression (13). With our findings, we confirm that PML plays tumor-promoting functions in TNBC, as its suppression delayed tumor growth in vivo, but we also identify PML as a regulator of metastasis by promoting the expression of a number of prometastatic genes regulated by HIF1A. Interestingly, as SOX9 is also regulated by HIF1A (45), our findings — along with those of Carracedo and collaborators (12, 13) — may converge into the identification of a novel HIF1A-PML axis that plays multiple oncogenic functions in TNBC.
Although, to our knowledge, this is the first experimental evidence that directly implicates PML in the regulation of metastasis, previous data has indicated that PML promotes cell migration, even in cell types where it exerts tumor-suppressive functions, such as mouse embryonic fibroblasts (46). Taken together, these studies indicate that the prometastatic function of PML may be more general and should be investigated in other tumor types where PML expression is maintained.
With our work, we position PML within an HIF1A-regulated network in TNBC by finding that PML is a HIF1A target gene whose expression is promoted by constitutive HIF1A activity. In apparent contrast with our results, it was described that HIF1A induces PML degradation in prostate cancer by promoting expression of the KLHL20 ubiquitin ligase (47). However, PML degradation reportedly occurred in conditions of mild hypoxia, while more severe hypoxia induced PML expression also in prostate cancer cells (47). Therefore, it appears that the outcome of PML regulation by HIF1A is complex and may depend on a number of factors, including levels or persistence of HIF1A activation, cell-type specificity, or normoxic versus hypoxic HIF1A expression. Within this line of reasoning, it is notable that TNBC cells display constitutive HIF1A expression through oxygen-independent mechanisms that are recently beginning to be elucidated (7, 16).
Upon being induced by HIF1A, PML cooperatively regulates a number of prometastatic HIF1A target genes in a tissue-specific manner in TNBC. These findings underline another layer of complexity in the functional interaction between PML and HIF1A. Indeed, besides the positive interaction herein described, PML was previously identified as an inhibitor of HIF1A accumulation in prostate cancer (14, 47), thus indicating that the cross-regulation of PML and HIF1A is complex and tumor-specific, and more work is necessary to identify the molecular determinants that finely regulate their functional interplay.
Our findings, along with those of Carracedo and collaborators (12, 13), may have important therapeutic implications. Taken together, they indicate that arsenic trioxide alleviates the metastatic features of aggressive breast cancers, while also impairing tumor growth and tumor-initiating capacity. In agreement with our data, previous studies have shown that arsenic trioxide exerts antitumor effects in breast cancer, and it inhibits cell migration and invasion in a TNBC cell line (48, 49). More importantly, it was recently reported that breast cancer mortality rates dropped in a region of northern Chile concomitantly with the presence of high concentrations of inorganic arsenic in drinking water and in stark contrast to increased incidence and mortality from a number of other solid tumors (50). We now provide an important basis for testing the efficacy of arsenic trioxide as an antitumor and antimetastatic agent for patients affected by TNBC or with high PML expression in their tumors. Our preclinical studies where arsenic trioxide was tested in collaboration with chemotherapy indicate that neo-adjuvant or adjuvant treatments that incorporate arsenic trioxide may block cancer reseeding, as well as the cancer-initiating capacity of residual cancer cells, and therefore may be proposed for future testing in clinical trials.
-
Methods
Cell culture, treatments, and reagents. NIH-3T3, HEK-293, MCF7, MDA-MB-361, BT-549, and MDA-MB-231 cells (ATCC) were maintained in DMEM; HEK-293T (ATCC) in IMDM; ZR-75 and 4T1 cells (ATCC) in RPMI; SUM-149 and SUM-159 (Asterand Biosciences) in Ham’s F12 (supplemented with 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 10 mM hepes). All media were from Lonza supplemented with 10% FBS (Euroclone) and 1% Pen-Strep antibiotics (Lonza). All cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2, except for SUM-149 and SUM-159, which were maintained at 10% CO2. For time course experiments with hypoxia mimetic agents, cells were treated with deferoxamine or cobalt chloride (Sigma-Aldrich) at a concentration of 100 μM for the indicated time. For in vitro treatment with arsenic trioxide, cells were incubated for 48 hours with 1 μM or 2 μM arsenic trioxide (Sigma-Aldrich).
Lentiviral vectors. GIPZ HIF1A shRNA or control shRNA plasmids were from Open Biosystems, while PML shRNAs or control shRNA plasmides were from the MISSION shRNA library (Sigma-Aldrich). Lentiviral vectors were obtained by HEK-293T transfection with calcium phosphate and subsequent concentration as previously described (51).
PML promoter cloning, luciferase reporter construction, and luciferase assays. The murine Pml promoter-luciferase construct was provided by Scott Lowe (Memorial Sloan Kettering Cancer Center, New York, USA; ref. 52). For human PML promoter, a 718-bp DNA fragment upstream of the transcription start site was amplified by PCR using the following primers: 5′-GGGGTACCCCATGCACAGCTGATCGTGTTGTTCC-3′ and 5′-CCGCTCGAGCGGTTGGAGTGCGTGAAGAGAAG-3′. The promoter fragment was then cloned into the PGL3-Basic vector (Promega). For luciferase activity assay NIH-3T3 or HEK-293 cells were plated in 24-well plates, and transient transfection was carried out 24 hours later using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. A plasmid containing a mutant, stable form of HIF1A (gifts of Celeste Simon, University of Pennsylvania, Philadelphia, USA) was cotransfected where indicated. When indicated, 16 hours after transfection, cells were treated with deferoxamine for the indicated time. Transfection normalization was obtained by cotransfecting a renilla expressing plasmid. Dual-Luciferase Reporter Assay System (Promega) was used to measure firefly and renilla luciferase activities and their ratio were calculated in a GloMax luminometer (Promega).
Quantitative PCR. Total RNA was isolated from cell lines using the RNeasy mini Kit (Qiagen). cDNA was synthesized by retrotranscription using Advantage RT-for-PCR Kit (Ambion). Real-time PCR (RT-PCR) was performed by TaqMan assay using a 7900 Fast-Real Time PCR System (Applied Biosystem). All probes for TaqMan assays were purchased from Applied Biosystem. As internal control, 18S was used. The relative fold-change expression of each mRNA was calculated using the 2-ΔΔCT method, except for assessing the relative expression of PML in human breast cancer cell lines where the 2-ΔCT was used.
Immunoblot. Proteins were extracted in the following buffer: 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.5% NP-40, supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific). After brief sonication, proteins were resolved by standard SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane through transBlot Turbo Transfer System (Bio-Rad). Membranes were incubated with the following antibodies: rabbit polyclonal HIF1A (Cayman, 10006421), rabbit polyclonal PML (Novus, NB10059787), and mouse β-actin (Santa Cruz Biotechnology Inc., sc-69879, AC-15). Secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology Inc.) were used, and immunoreactive proteins were detected using the ECL Western Blotting Detection Reagents (GE Healthcare).
ChIP assay. ChIP experiments were performed as previously described (53). Protein-DNA fragments complexes were immunoprecipitated either with monoclonal HIF1A antibody (Novus NB100-105, H1alpha67) or monoclonal PML antibody (Santa Cruz Biotechnology Inc., sc-966, PG-M3), or normal mouse IgG (Santa Cruz Biotechnology Inc., sc-2025) as control. To amplify DNA fragments obtained by ChIP in the regions of interest (Figure 1F and Figure 3, D and E) primer sets (5′-3′) used are listed in Table 1.
Migration assay. For migration assay, cells were seeded and allowed to reach confluence. Wounds (at least 3 per plate) were made by cell scraping with a 1-ml tip. After 3 washes with sterile PBS to remove scraped-off cells, images of the wounds were taken in bright field microscopy and cells were incubated for 24 hours at 5% CO2, except for SUM-159, which were incubated for 36 hours at 10% CO2. For experiments with arsenic trioxide, wound scraping was performed 24 hours after addition of the compound, and fresh arsenic was added after PBS washes at the same doses. Images of the same wound spots were taken at the end of the incubation, and wound areas were quantified through ImageJ software. Wound areas at the end of the experiment were divided by wound areas calculated at the beginning of the experiment, in triplicate. The average values of technical triplicates thus obtained upon PML silencing or treatment with arsenic trioxide were divided by average values obtained from control cells; the reciprocal number of the obtained value was represented in figures.
Invasion assay. For invasion assays, 100,000 cells were seeded in duplicate on Matrigel-coated transwells (BD Biosciences, Millipore). The lower chambers were filled with NIH 3T3-conditioned medium as previously described (54). After 5 hours, cells migrating to the lower side of the transwell were stained with crystal violet. Cells were counted in the images acquired from at least 3 fields (20×) per experimental point, in duplicate. Values represented in the figures represent the average of biological replicates, as indicated in figure legends.
Cell viability and proliferation. Cell viability was assessed by trypan blue or annexin V/7-AAD staining. To assess cytotoxic and cytostatic effect of treatment with arsenic trioxide, 40 × 103 cells were seeded in 12-well plates and treated with 1 μM or 2 μM arsenic trioxide for 48 hours. Cell numbers and cell viability was then evaluated by Trypan Blue. For cell proliferation, 20 × 103 cells were seeded in 12-well plates and counted every 24 hours.
Mouse models. For breast cancer xenograft experiments, 2 × 106 MDA-MB-231 cells resuspended in matrigel/PBS (1:3, 45 μl total) were injected in the mammary fat pad of 6- to 8-week old NOD scid γ (NSG) immunocompromised mice (Charles River Laboratories). For allograft experiments, 2 × 106 4T1 cells resuspended in matrigel/PBS (1:3, 45 μl total) were injected in the mammary fat pad of 6- to 8-week-old BALB/c mice (Charles River Laboratories). Tumor growth was measured by caliper with the (W2 × L)/2 formula. For in vivo treatment with arsenic trioxide, mice were treated with 8 mg/kg or 4 mg/kg arsenic trioxide as indicated by i.p. injection 5 days a week starting when tumors became palpable. Where indicated, mice were treated with paclitaxel 20 mg/Kg twice a week by i.p. injection (41) Mice were euthanized when primary tumors reached similar sizes in the cohorts that were being compared, and the lungs were perfused with PBS before excision. Lungs and primary tumors were fixed in formalin, and sections were subjected to H&E staining and immunohistochemical analysis. For PML and HIF1A IHC, the following antibodies and dilutions were used: monoclonal PML antibody from DAKO (PG-M3, 1: 200) and rabbit monoclonal HIF1A antibody from Epitomics (ab51608, EP1215Y, 1: 800).
Microarray data analysis. Sample classification into high- and low-hypoxia response tumors was based on the expression level of 123 “epithelial hypoxic signature” genes in 295 early-stage breast cancer samples accrued and analyzed by the Netherlands Cancer Institute (17). Expression profiles at the gene level for 547 breast cancer samples, assayed with Agilent microarrays, were retrieved from the TCGA Data Portal website (https://tcga-data.nci.nih.gov/docs/publications/brca_2012/). Profiles for genes belonging to the hypoxia signature proposed by Chi et al. (17) were selected (only 119 genes could be mapped) and processed through average linkage hierarchical clustering based on the Pearson’s correlation coefficient metric on both genes and samples. Cluster enrichment for the TNBC was assessed by means of the Fisher’s exact test. Significance of the differential expression of PML between the identified cluster and all other breast cancer samples was computed using a Student’s 2-tailed t test. Genes belonging to the hypoxia signature that were found upregulated in TNBC samples compared with non-TNBCs were ranked according to their significance determined by applying a False Discovery Rate (FDR) corrected Student’s 2 tailed t test. Linear correlation between the expression levels of PML and the other genes was computed as Pearson’s correlation coefficient (r) provided with the correspondent statistical significance, testing the null hypothesis that the coefficient is zero. The analyses were accomplished in the Matlab (The MathWorks Inc.) environment.
Recurrence analysis. Follow-up data of 383 TNBC samples and their gene expression profiles (NCBI GEO GSE31519) (36) were exploited to derive the Kaplan-Meier curves for event-free survival, where the endpoint was local and distant recurrence. PML expression values were dichotomized at the median and significance was computed by applying the log rank test; analyses were performed using the R (http://www.r-project.org/) library “survival.”
Statistics. Student’s 2-tailed t test was used to determine statistical significance between 2 groups. For multiple comparison analysis, statistical analysis was performed using 1-way ANOVA followed by Tukey’s post-hoc multiple comparison test. The significance level was set at P < 0.05.
Study approval. All the animals used in this study were maintained in pathogen-free animal facility and treated in accordance with European Union guidelines; animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of IRCCS San Raffaele Scientific Institute.
-
Author contributions
MP performed the majority of the experiments and cowrote the manuscript. LC performed gene expression analysis and migration assays in TNBC cell lines. RC performed PML expression analysis and luciferase assays in NIH-3T3 and HEK-293 cells. AP cloned the human PML promoter in the luciferase-expressing vector and performed PML expression analysis with hypoxia-mimetic agents. GAD tested the effect of arsenic trioxide in vitro in breast cancer cell lines. NC analyzed HIF1A and PML expression. RV performed HIF1A target gene expression analysis in cell lines. AV and UC expanded and characterized SUM-159 cells. LP performed all the bioinformatics analysis on breast cancer patients. CD is a certified pathologist who analyzed mouse tissues. RB designed the study, supervised the research, and cowrote the manuscript.
-
Acknowledgments
The authors are grateful to all present and past members of the laboratory of RB for collective thinking and support; Martina Rocchi and Francesca Invernizzi for assistance in processing and IHC of mouse tissues; Luca Gianni, Milvia Zambetti, Stefania Zambelli, and Giampaolo Bianchini of the Department of Oncology of San Raffaele Hospital for support and discussion; the laboratories of Ivan de Curtis, Davide Gabellini, and Vincenzo Russo for sharing protocols and reagents; Elisa de Stanchina and Scott Lowe for providing the mouse PML promoter-luciferase construct; Celeste Simon for providing a plasmid containing the stable form of HIF1A; and Giliola Calori for support with statistical analysis. This work was financially supported by the Giovanni Armenise-Harvard Foundation with a career development award to RB and by Ospedale San Raffaele with an OSR Pilot and Seed grant to RB.
Address correspondence to: Rosa Bernardi, Division of Experimental Oncology, IRCCS San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy. Phone: 39.02.26435606; E-mail: bernardi.rosa@hsr.it.
-
Footnotes
RC’s present address is: IFOM, the FIRC Institute of Molecular Oncology, Milan, Italy.
AP’s present address is: Department of Biochemistry and Molecular Genetics, Feinberg School of Medicine, Northwestern University, Chicago, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(4):e87380. https://doi.org/10.1172/jci.insight.87380.
-
References
- Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19(11):1450–1464.
- Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–218.
- Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer--deciphering the heterogeneity. Clin Cancer Res. 2014;20(4):782–790.
- Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70.
- Montagner M, et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487(7407):380–384.
- Chen X, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508(7494):103–107.
- Lin A, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–224.
- Semenza GL. Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends Mol Med. 2012;18(9):534–543.
- Liu ZJ, Semenza GL, Zhang HF. Hypoxia-inducible factor 1 and breast cancer metastasis. J Zhejiang Univ Sci B. 2015;16(1):32–43.
- Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430–439.
- Mazza M, Pelicci PG. Is PML a Tumor Suppressor? Front Oncol. 2013;3:174. View this article via: PubMed Google Scholar
- Carracedo A, et al. A metabolic prosurvival role for PML in breast cancer. J Clin Invest. 2012;122(9):3088–3100.
- Martín-Martín N, et al. Stratification and therapeutic potential of PML in metastatic breast cancer. Nat Commun. 2016;7:12595. View this article via: PubMed Google Scholar
- Bernardi R, et al. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442(7104):779–785.
- Bernardi R, et al. Pml represses tumour progression through inhibition of mTOR. EMBO Mol Med. 2011;3(5):249–257.
- Briggs KJ, et al. Paracrine Induction of HIF by Glutamate in Breast Cancer: EglN1 Senses Cysteine. Cell. 2016;166(1):126–139.
- Chi JT, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3(3):e47.
- Erler JT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–1226.
- Gilkes DM, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73(11):3285–3296.
- Dupuy F, et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015;22(4):577–589.
- McDonald PC, Winum JY, Supuran CT, Dedhar S. Recent developments in targeting carbonic anhydrase IX for cancer therapeutics. Oncotarget. 2012;3(1):84–97.View this article via: PubMed Google Scholar
- Zhang J, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72(17):4597–4608.
- Cronin PA, Wang JH, Redmond HP. Hypoxia increases the metastatic ability of breast cancer cells via upregulation of CXCR4. BMC Cancer. 2010;10:225. View this article via: PubMed Google Scholar
- Siclari VA, et al. Tumor-expressed adrenomedullin accelerates breast cancer bone metastasis. Breast Cancer Res. 2014;16(6):458.
- Zhang H, et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31(14):1757–1770.
- Blouin MJ, et al. Germ line knockout of IGFBP-3 reveals influences of the gene on mammary gland neoplasia. Breast Cancer Res Treat. 2015;149(3):577–585.
- Wang D, et al. Oct-4 and Nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget. 2014;5(21):10803–10815.
- Kho DH, et al. Autocrine motility factor modulates EGF-mediated invasion signaling. Cancer Res. 2014;74(8):2229–2237.
- Wang J, et al. A glycolytic mechanism regulating an angiogenic switch in prostate cancer. Cancer Res. 2007;67(1):149–159.
- Hu Z, et al. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med. 2009;7:9. View this article via: PubMed Google Scholar
- Bracken CP, et al. Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. EMBO J. 2014;33(18):2040–2056.
- Draheim KM, Chen HB, Tao Q, Moore N, Roche M, Lyle S. ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene. 2010;29(36):5032–5047.
- Antón IM, Jones GE, Wandosell F, Geha R, Ramesh N. WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol. 2007;17(11):555–562.
- Staub E, et al. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. J Mol Med. 2009;87(6):633–644.
- Si W, et al. Dysfunction of the Reciprocal Feedback Loop between GATA3- and ZEB2-Nucleated Repression Programs Contributes to Breast Cancer Metastasis. Cancer Cell. 2015;27(6):822–836.
- Rody A, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13(5):R97.
- Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2(5):E85–E90.
- Kumar PP, et al. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9(1):45–56.
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74.
- de Thé H, Le Bras M, Lallemand-Breitenbach V. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198(1):11–21.
- Enriquez-Navas PM, et al. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Transl Med. 2016;8(327):327ra24.
- Martin-Martin N, Sutherland JD, Carracedo A. PML: Not all about Tumor Suppression. Front Oncol. 2013;3:200. View this article via: PubMed Google Scholar
- Ito K, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453(7198):1072–1078.
- Ito K, et al. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350–1358.
- Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134(21):3917–3928.
- Tang MK, et al. Promyelocytic leukemia (PML) protein plays important roles in regulating cell adhesion, morphology, proliferation and migration. PLoS One. 2013;8(3):e59477.
- Yuan WC, et al. A Cullin3-KLHL20 Ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell. 2011;20(2):214–228.
- Chow SK, Chan JY, Fung KP. Inhibition of cell proliferation and the action mechanisms of arsenic trioxide (As2O3) on human breast cancer cells. J Cell Biochem. 2004;93(1):173–187.
- Ahn RW, et al. A novel nanoparticulate formulation of arsenic trioxide with enhanced therapeutic efficacy in a murine model of breast cancer. Clin Cancer Res. 2010;16(14):3607–3617.
- Smith AH, et al. Rapid reduction in breast cancer mortality with inorganic arsenic in drinking water. EBioMedicine. 2014;1(1):58–63.
- Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25(2):217–222.
- de Stanchina E, et al. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13(4):523–535.
- Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831.
- Shaw LM. Tumor cell invasion assays. Methods Mol Biol. 2005;294:97–105.View this article via: PubMed Google Scholar
-
Version history
-
Version
1
(February 23, 2017):
Electronic publication
-
Version
1
(February 23, 2017):









