Research ArticleMetabolismNephrology
Open Access |  10.1172/jci.insight.189130
10.1172/jci.insight.189130
NSD1-916aa encoded by CircNSD1 contributes to AKI-to-CKD transition through inducing ferroptosis in tubular epithelial cells
Li Gao,1,2,3 Junsheng Zhang,4,5 Chaoyi Chen,2 Sai Zhu,1,2 Xianglong Wei,2 Guiqin Tang,2 Sheng Wang,3 Yukai Wang,2 Xinran Liu,2 Ling Jiang,2 and Yonggui Wu2,3
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Gao, L. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Zhang, J. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Chen, C. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Zhu, S. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Wei, X. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Tang, G. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Wang, S. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Wang, Y. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Liu, X. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Jiang, L. in: PubMed | Google Scholar
1Inflammation and Immune-Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, and
2Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
3Center for Scientific Research of Anhui Medical University, Hefei, China.
4Clinical Pathology Center, The First Affiliated Hospital of Anhui Medical University, Hefei, China.
5Anhui Public Health Clinical Center, Hefei, China.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
Find articles by Wu, Y. in: PubMed | Google Scholar
Published July 15, 2025 - More info
JCI Insight. 2025;10(16):e189130. https://doi.org/10.1172/jci.insight.189130.
© 2025 Gao et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: November 11, 2024; Accepted: July 7, 2025
-
Abstract
Acute kidney injury (AKI) is characterized by a rapid decline in renal function. In severe or recurrent cases, AKI can progress to chronic kidney disease (CKD), marked by renal inflammation and fibrosis. Despite the severity of these outcomes, early-stage diagnostic tools and pharmacological interventions for AKI-to-CKD progression remain limited. In this study, we examined circular RNA (circRNA) expression profiles in mouse renal cortex tissues 14 days after ischemia/reperfusion (I/R) injury using circRNA-Seq. The renal biopsy samples of patients after AKI exhibited reduced CircNSD1 expression, which was inversely associated with inflammation and fibrosis. Overexpression of CircNSD1 attenuated ferroptosis in vivo and in vitro, while slowing AKI-to-CKD progression. Mechanistically, CircNSD1 downregulated ACSL4 and SLC39A14 expression through histone H3 lysine 36 (H3K36) methylation, a critical pathway regulating ferroptosis after AKI or hypoxia/reoxygenation (H/R) injury. Furthermore, we identified that CircNSD1 encoded a NSD1-916aa peptide, which may functionally contribute to its observed effect. Collectively, these findings demonstrated that CircNSD1 may serve as a diagnostic and therapeutic target for early detection of AKI-to-CKD transition.
-
Introduction
Acute kidney injury (AKI) is a critical clinical condition characterized by a sudden and pronounced deterioration in kidney function, often marked by proximal tubules damage and elevated serum creatinine and blood urea nitrogen (BUN) levels, and is associated with substantial morbidity and mortality rates (1). Furthermore, a considerable number of individuals with AKI experience progressive inflammation and fibrosis, ultimately developing chronic kidney disease (CKD) (2). Statistical analyses have indicated that patients with AKI are at a 2.67-fold higher risk of CKD and a 4.81-fold higher risk of end-stage renal disease (3, 4). Despite the severity of these outcomes, there is a lack of effective diagnostic tools and pharmacological interventions to treat AKI or mitigate its progression to CKD. Additionally, a comprehensive understanding of the molecular mechanisms underlying the AKI-to-CKD transition has yet to be fully elucidated, posing a challenge to the development of targeted clinical strategies (5).
Failed repair of renal epithelial tubular cells (PT cells) leads to a transition to a distinct proinflammatory state, which is implicated in the progression of fibrosis after AKI. The downregulation of genes involved in glutathione metabolism renders PT cells more susceptible to ferroptotic stress, intensifying the inflammatory response. The induction of high ferroptotic stress through genetic manipulation after injury aggravates inflammation and fibrosis, which are key drivers of AKI-to-CKD transition (6). Ferroptotic stress is characterized by an excessive accumulation of lipid reactive oxygen species, disruption in the metabolism of polyunsaturated fatty acids, and damage to mitochondrial function. This process participates in PT cell repair (6, 7). Emerging evidence highlights the protective effects of small-molecule inhibitors of ferroptosis, such as ferrostatin-1 (Fer-1) against AKI (8). However, the pathogenic mechanisms through which ferroptosis influences the transition from AKI to CKD remain poorly defined.
Advancements in next-generation sequencing have identified a multitude of uncharacterized transcripts. Among these, noncoding RNAs (ncRNAs) that lack an open reading frame (ORF) are traditionally considered nontranslatable. However, exonic circular RNAs (circRNAs) can harbor complete or partial ORFs corresponding to their linear parental genes, suggesting that certain circRNAs previously classified as noncoding may, in fact, be translatable. For instance, Circ-ZNF609 encoded ZNF609-250aa to induce cell apoptosis and AKI by impairing the autophagy flux via AKT/mTOR-dependent mechanism (9). Similarly, circular MTHFD2L RNA-encoded CM-248aa inhibits gastric cancer progression by targeting the SET-PP2A interaction (10). In glioblastoma, a circular RNA (circMET, hsa_circ_0082002)-encoded MET variant has been shown to drive tumorigenesis (11). Despite growing insights into the roles of protein-coding circRNAs in various pathological contexts, their contributions to renal disease, particularly during the transition from AKI to CKD, remain poorly understood.
In this study, we identified a circRNA, designated CircNSD1, through high-throughput sequencing analysis of renal cortex tissues harvested from mice 14 days following ischemia/reperfusion (I/R) injury. CircNSD1 expression was markedly downregulated in renal tissues from AKI-to-CKD mouse models, as well as in patient samples and hypoxia/reoxygenation–induced (H/R-induced) HK-2 cells. To elucidate the role of CircNSD1 in the transition from AKI to CKD, we overexpressed CircNSD1 both in vivo and in vitro. Subsequently, we conducted an untargeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) lipidomics analysis to compare H/R-treated control cells with those overexpressing CircNSD1, aiming to uncover the molecular mechanisms. Furthermore, we explored the potential of CircNSD1 to encode a bioactive peptide. Collectively, our findings suggest that CircNSD1 may serve as a promising biomarker and therapeutic target for early intervention in the AKI-to-CKD transition.
-
Results
Injured epithelia drive inflammation and fibrosis contribution to AKI-to-CKD transition. To establish the mouse model of AKI-to-CKD transition, we analyzed a single-cell RNA-Seq dataset of mouse kidney published in PNAS in 2020 (GSE139107) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139107). As shown in Figure 1A, cell clusters named NewPT1 and NewPT2 in PNAS (7), positive for the injury marker Havcr1, exhibited sustained high expression of inflammation-related genes (Ccl2, Cxcl1, Cxcl2, and Cxcl10) and fibrosis-related genes (Tgfb1, Tgfb2 and Tgfbr2) from 2 days to 6 weeks after I/R. Figure 1B illustrates the AKI-to-CKD transition mouse model at various time points following I/R: 0 days, 2 days, 7 days, 14 days, and 6 weeks. As anticipated, serum creatinine, estimated glomerular filtration rate (GFR), H&E, and periodic acid–Schiff (PAS) staining revealed extensive tubular epithelial injury and rapid renal function decline at 2 days after I/R. Partial functional recovery was observed from 7 days to 6 weeks (Figure 1, C–E, and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.189130DS1). Meanwhile, macrophage infiltration (F4/80), fibrotic deposition (Masson’s trichrome), and α–smooth muscle actin-marked (ASMA-marked) fibrotic areas were milder on day 7 but intensified by day 14 and persisted through week 6 (Figure 1, F–H, and Supplemental Figure 1B). These findings indicate that injured epithelial cells drive renal inflammation and fibrosis, which contributed to AKI-to-CKD transition. The subsequent inflammation and fibrosis mediated by injured epithelial cells can be observed substantially, suggesting that day 14 of ischemia is the optimal time point for AKI-to-CKD model.
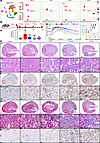 Figure 1
Figure 1Fourteen-day I/R as a model for AKI-to-CKD transition in mice. (A) Reanalyzing a single-cell RNA-Seq dataset of mouse kidney (GSE139107) in KIT websites. (B) An illustration of AKI-to-CKD transition mouse model. (C) Serum creatinine levels. (D) GFR. (E) H&E staining. (F) IHC staining for F4/80. (G) Masson’s trichrome staining. (H) IHC staining for ASMA. n = 6–7. Scale bar: 100 μm (red), 50 μm (black). Data are shown as mean ± SEM, ***P < 0.001 compared with the control by 1-way ANOVA with Tukey’s post hoc test. ###P < 0.001 compared with the I/R-2D group by 1-way ANOVA with Tukey’s post hoc test.
CircNsd1 expression is suppressed in mouse model of AKI-to-CKD transition, in H/R-induced epithelial cells and patients after AKI. To elucidate the role of circRNAs in the AKI-to-CKD transition, the renal cortex tissues from mice 14 days after I/R were subjected to high-throughput circRNA-Seq. Heatmap demonstrated the top 7 differentially expressed known circRNAs (Figure 2A). Subsequent validation via real-time PCR from 2 days to 6 weeks after I/R excluded circRNAs with insignificant expression changes or low homology during transition. CircNsd1 emerged as the most dynamically altered circRNA with the highest basal expression (Figure 2B). FISH assay revealed that CircNsd1 predominantly localized in the cytoplasm of tubular epithelial cells, with its expression decreasing in mice following I/R injury or cisplatin injury (Figure 2C and Supplemental Figure 1, C and D).
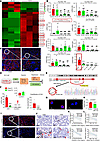 Figure 2
Figure 2CircNSD1 expression is suppressed in vivo, in vitro, and in patients. (A) Results of high-throughput circRNA-Seq in kidney tissues from control and I/R-14D mice. (B) Relative expression of circRNA detected by real time-PCR from 2 days to 6 weeks after I/R injury. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001 compared with the control or CircNsd1 group by 1-way ANOVA with Tukey’s post hoc test. #P < 0.05, ##P < 0.01, ###P < 0.001 compared with the I/R-2D group by 1-way ANOVA with Tukey’s post hoc test. (C) FISH assay for CircNSD1. Scale bar: 100 μm (red), 50 μm (white). (D) An illustration of H/R-induced HK-2 cells model. (E) Examination of CircNSD1 in circBase (http://circbase.org/) via Sanger sequencing. (F) Real-time PCR analysis of CircNSD1 and parental gene NSD1, digested by RNase R. Data are shown as mean ± SEM, ***P < 0.001 compared with RNase R- RF1 group by 1-way ANOVA with Tukey’s post hoc test. (G) FISH assay for CircNSD1 in HK-2 cells and H/R-induced HK-2 cells. Scale bar: 20 μm (white). (H) CircNSD1 was detected in serum of control group and patients after AKI. Data are shown as mean ± SEM, ***P < 0.001 compared with Control group by Student’s t test. (I) FISH for CircNSD1 in renal biopsies from controls and patients after AKI. Scale bar: 100 μm (white); 50 μm (black). (J) IHC for CD68+ reveals the inflammatory response, and ASMA reveals fibrosis in renal biopsies from controls and patients with AKI-to-CKD. Scale bar: 100 μm (black). (K) Correlation analysis between CircNSD1 in renal biopsies and the inflammatory response (CD68+) or fibrosis (ASMA) in patients after AKI. (L) Correlation analysis between CircNSD1 in serum and the inflammatory response (CD68+) or fibrosis (ASMA) in renal biopsies from patients after AKI.
Cellular models utilizing HK-2 cells were established to further validate the expression of circRNAs that exhibited marked changes (Supplemental Figure 1E and Figure 2D). Notably, CircNSD1 was confirmed as the circRNA with the most marked expression change and the highest level of basal expression level in vitro (Supplemental Figure 1F). CircNSD1, derived from the NSD1 gene, is located on chromosome 5 (positions 176618884–176639196) and comprises exons 3–5. As depicted in Figure 2E, Sanger sequencing confirmed the presence of a head-to-tail splice junction characteristic of circRNAs, which matched the reference sequence for CircNSD1 listed in circBase (http://circbase.org/). Considering that head-to-tail splicing products may be derived from genomic rearrangements or trans-splicing, the stability of CircNSD1 was assessed by its resistance to digestion by RNase R (Figure 2F), a highly processive 3′–5′ exonuclease. The results indicated that the expression level of CircNSD1 remained largely unaffected by RNase R treatment, whereas the expression of parental gene NSD1 was substantially reduced. FISH corroborated CircNSD1’s cytoplasmic localization in HK-2 cells (Figure 2G). Furthermore, compared with normal controls, the expression of CircNSD1 was decreased in serum and renal biopsies from patients after AKI (Figure 2, H and I). CircNSD1 expression levels in renal tissues of patients after AKI were substantially reduced and showed a negative correlation with both renal inflammatory and fibrotic markers. Meanwhile, serum circNSD1 levels were negatively correlated with inflammatory markers but showed no association with fibrosis — an observation that warrants further investigation with a large sample size (Figure 2, J–L).
Overexpression of CircNSD1 resists AKI-to-CKD transition in vivo and in vitro. To investigate the role of CircNSD1 in AKI-to-CKD transition, HK-2 cells were transfected with a lentiviral vector encoding CircNSD1. FISH assay revealed that CircNSD1 was overexpressed in the cytoplasm of tubular epithelial cells (Figure 3A). Real-time PCR further confirmed CircNSD1 overexpression in HK-2 (Figure 3B). Overexpression of circNSD1 markedly reduced ASMA protein staining in HK-2 cells subjected to H/R injury (Figure 3C). Real-time PCR further confirmed decreased expression of inflammation-related genes (TNFA, CCL2, IL6) and fibrosis-related genes (TGFB1, COL1, ASMA) in circNSD1-overexpressed HK-2 cells, compared with control HK-2 cells after H/R injury (Figure 3D)
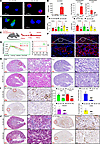 Figure 3
Figure 3CircNSD1 overexpression inhibits AKI-to-CKD transition. (A) FISH assay in CircNSD1 overexpression cells. Scale bar: 20 μm (white). (B) Real-time PCR analysis of CircNSD1 expression in CircNSD1 overexpression cells. (C) Immunofluorescence for ASMA in CircNSD1 overexpression cells. (D) Real-time PCR analysis of proinflammatory cytokines (TNFA, CCL2, and IL6) and fibrogenic genes (TGFB1, COL1, and ASMA) in H/R-48h induced control and CircNSD1 overexpression cells. (E) Schematic illustrating the approach of overexpressing CircNsd1 in mice. (F) FISH assay of CircNsd1 expression in kidney tissues from control and CircNsd1-overexpressing I/R mice. Scale bar: 100 μm (red), 50 μm (white). (G) GFR in control and CircNsd1-overexpressing I/R mice. (H) Representative H&E staining and PAS staining pictures in kidney tissues from control and CircNsd1-overexpressing I/R mice. (I) IHC staining of F4/80+ macrophages in kidney tissues from control and CircNsd1-overexpressing I/R mice. (J) Representative Masson and IHC staining of ASMA pictures in kidney tissues from control and CircNsd1-overexpressing I/R mice. Scale bar: 500 μm (red), 50 μm (black). Data are shown as mean ± SEM, #P < 0.05, ##P < 0.01, ###P < 0.001 compared with the EV-H/R-48h group or EV-I/R-14D group by Student’s t-test.
The role of CircNsd1 was further validated in a murine model, and systemic CircNsd1 overexpression was achieved in mice via tail vein injection of HBAAV2/9-mmu_circ_0000465 (CircNsd1-OE; Figure 3E). Exogenous overexpression of CircNsd1 in renal tissue was confirmed by real-time PCR and FISH (Figure 3F and Supplemental Figure 1G). PAS staining revealed no renal structural changes in sham-EV and sham-OE groups, confirming administration of HBAAV2/9-circNsd1 via tail vein injection had no effect on normal renal structure (Supplemental Figure 1H). At 14 days after I/R injury, CircNsd1 overexpression group (OE-I/R-14D) markedly improved GFR compared with EV controls (EV-I/R-14D) (Figure 3G). H&E and PAS staining further corroborated reduced renal injury in the OE-I/R-14D group (Figure 3H). IHC showed diminished F4/80+ macrophage infiltration in renal epithelia of OE-I/R-14D mice (Figure 3I). Masson’s trichrome staining and ASMA IHC confirmed that CircNsd1 overexpression substantially protected against I/R-induced AKI-to-CKD transition (Figure 3J). Collectively, these findings support that CircNsd1 overexpression protected against I/R-induced renal dysfunction, inflammation, and fibrosis, thereby impeding the transition from AKI-to-CKD.
Overexpression of CircNSD1 reduced ferroptosis in H/R-induced HK-2 cells and AKI-to-CKD mice. To decipher the underlying molecular mechanisms, untargeted LC-MS/MS lipidomics analysis of H/R-treated control and CircNSD1 overexpression cells was performed. Orthogonal partial least squares discriminant analysis (OPLS-DA) identified 193 differentially abundant metabolites (variable importance in projection [VIP] ≥ 1, P < 0.05). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed marked enrichment of ferroptosis-associated pathways, including ferroptosis and glutathione metabolism. Key ferroptosis-related metabolites included Glutathione, L-Glutamate, PC [18:1(11Z)/16:0], (±)5,6-DiHETrE, γ-Glu-Cys, and PC [22:0/18:4 (6Z,9Z,12Z,15Z)] (Figure 4A). Furthermore, CircNSD1 overexpression in H/R-treated cells substantially restored levels of ferroptosis markers (Fe²+, malondialdehyde [MDA], lipid peroxidation [LPO], and glutathione [GSH]) compared with controls (Figure 4B). Electron microscopy, DCF assay, and DHE assay corroborated that overexpression of CircNSD1 reduced ferroptosis under H/R condition (Figure 4C).
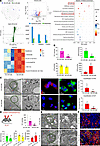 Figure 4
Figure 4CircNSD1 inhibits ferroptosis in AKI-to-CKD in vivo and in vitro. (A) Results LC-MS/MS lipidomics analysis of H/R-treated control and CircNSD1 overexpression cells. (B) Levels of ferroptosis-related metabolites, including Fe2+, MDA, LPO, and GSH in vitro. (C) Electron microscopy, DCF assay and DHE assay in H/R-treated CircNSD1 overexpression cells and controls. Scale bar: 2 μm (left); 500 nm (right); 20 μm (DHE and DCF staining). (D) An illustration of CircNsd1 overexpression in mice model and metabolic (Fe2+, MDA, LPO and GSH) analysis in I/R-induced control and CircNsd1 overexpression mice. (E) Representative electron micrographs of in kidney tissues from control and CircNsd1-overexpressing I/R mice. Scale bar: 2 μm (left); 500 nm (right). (F) Representative staining GPX4 in kidney tissues from control and CircNsd1-overexpressing I/R mice. Scale bar: 50 μm. Data are shown as mean ± SEM, #P < 0.05, ##P < 0.01, ###P < 0.001, compared with EV-H/R-48h group or EV-I/R-14D by Student’s t test.
To validate these findings in vivo, we assessed ferroptosis-associated metabolites and proteins in I/R-induced control and CircNsd1 overexpressing mice. Metabolic (Fe2+, MDA, LPO, and GSH) analysis, electron microscopy, and GPX4 IF assay showed that CircNsd1 overexpression mitigated ferroptosis in AKI-to-CKD mice (Figure 4, D–F). These results support that overexpression of CircNSD1 alleviated ferroptosis in H/R-induced HK-2 cells and AKI-to-CKD mice.
Overexpression of CircNSD1 decreased Erastin-induced ferroptosis, inflammation, and fibrosis in HK-2 cells. To investigate whether the role of CircNSD1 in inflammation and fibrosis via ferroptosis pathway, control and CircNSD1-overexpressing cells were treated with the ferroptosis inducer Erastin. Real-time PCR indicated that treatment of HK-2 cells with 5 μM Erastin for 48 hours led to increased expression of inflammation-related genes (TNFA, CCL2, and IL6) and fibrosis-related genes (TGFB1, COL1, and ASMA) (Figure 5, A and B). Subsequent assays, including electron microscopy; metabolic analysis of Fe2+, MDA, LPO, and GSH level; and DHE/DCF fluorescence assay, collectively suggested that overexpression of CircNSD1 marked reduced Erastin-induced ferroptosis (Figure 5, C–E). Concurrently, real-time PCR showed a decrease in the expression of inflammation-related genes (TNFA, CCL2, and IL6) and fibrosis-related genes (TGFB1, COL1, and ASMA) in CircNSD1-overexpressing cells following Erastin treatment (Figure 5, F and G). Furthermore, IF staining of ASMA further confirmed a diminished fibrotic response in the overexpression group compared with the control group (Figure 5H). Collectively, these findings support that overexpression of CircNSD1 alleviated inflammation and fibrosis induced by ferroptosis in vitro.
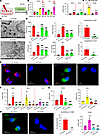 Figure 5
Figure 5Overexpression of CircNSD1 decreases ferroptosis, inflammation, and fibrosis in Erastin-induced HK-2 cells. (A) An illustration of Erastin-induced HK-2 cells model. (B) Real-time PCR analysis of inflammation-related genes (TNFA, CCL2, and IL6) and fibrosis-related genes (TGFB1, COL1, and ASMA) in HK-2 cells after 48 hours of Erastin induction. (C) Representative electron micrographs of control and CircNSD1-overexpressing cells induced by Erastin. Scale bar: 500nm. (D)Levels of ferroptosis-related metabolites,including Fe2+, MDA, LPO and GSH in cells. (E) DCF assay and DHE assay in Erastin-induced CircNSD1 overexpressing cells and controls. Scale bar: 20μm. (F) Real-time PCR analysis of proinflammatory cytokines (TNFA, CCL2 and IL6) in control and CircNSD1 overexpressing cells after 48 hours of Erastin induction. (G) Real-time PCR analysis of fibrosis-related genes (TGFB1,COL1 and ASMA) in Erastin-treated control and CircNSD1 overexpressing cells. (H) Representative immunofluorescence staining of ASMA pictures in control and CircNSD1 overexpressing cells treated with Erastin. Scale bar: 20 μm. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001 compared with the control by 1-way ANOVA with Tukey’s posthoc test. #P < 0.05, ##P < 0.01, ###P < 0.001 compared with the EV-Erastain group by 1-way ANOVA with Tukey’s post hoc test.
Overexpression of CircNSD1 downregulated ACSL4 and SLC39A14 by H3K36 methylation. Ferroptosis, a nonapoptotic form of cell death, plays a critical role in the AKI-to-CKD transition (12). During this transition, key ferroptosis-related proteins such as GPX4 and ACSL4 exhibit dynamic changes. Immunofluorescence, real-time PCR, and spatial transcriptomics datasets (GSE139107) reveal decreased GPX4 expression in early AKI stages, with partial restoration during CKD progression, alongside persistently elevated ACSL4 levels in renal epithelial cells (Supplemental Figure 1, I and J). To examine the direct mechanisms by which CircNsd1 regulates ferroptosis, reanalysis of single-cell datasets (GSE139107) was performed to identify differentially expressed genes (log2FC > 0.26 and P < 0.05) at 2 and 14 days after AKI compared with the control group (Figure 6A). Intersection with FerrDb (a ferroptosis database; http://www.zhounan.org/ferrdb/current/) ferroptosis-related genes yielded 26 candidates. KEGG pathway analysis of the resulting 26 genes revealed that Acsl4, Lpcat3, Slc39a14, and Sat1 are involved in the ferroptosis pathway (Figure 6B). Therefore, we focused on investigating the regulation of Acsl4, Lpcat3, Slc39a14, and Sat1 by CircNsd1.
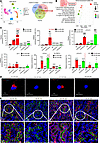 Figure 6
Figure 6Overexpression of CircNSD1 downregulated ACSL4 and SLC39A14 by H3K36 methylation. (A) Reanalyzing a single-cell RNA-Seq dataset of mouse kidney (GSE139107). Venn diagram showing the intersection analysis of genes changed in I/R-2D and I/R-14D with genes associated with ferroptosis. (B) KEGG pathway analysis of overlapping genes. (C) ChIP assay detecting the H3K36me modification of ACSL4, LPCAT3, SLC39A14, and SAT1 regulated by CircNSD1 in control and CircNSD1 overexpressed cells treated with H/R-48h. (D) Representative immunofluorescence staining of ACSL4 and SLC39A14 in control and CircNSD1 overexpressed cells treated with H/R-48h. Scale bar: 20 μm. (E) Immunofluorescence staining of ACSL4 and LTL in kidney tissues from control and CircNsd1-overexpressing I/R mice. Immunofluorescence staining of SLC39A14 and LTL in kidney tissues from control and CircNsd1-overexpressing I/R mice. Scale bar: 50 μm. Data are shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001 compared with the IgG-EV-H/R-48h group by 1-way ANOVA with Tukey’s post hoc test. ###P < 0.001 compared with H3K36me1-EV-H/R-48h group or H3K36me2-EV-H/R-48h group by 1-way ANOVA with Tukey’s post hoc test. &&&P < 0.001 compared with H3K36me3-EV-H/R-48h group by 1-way ANOVA with Tukey’s post hoc test.
The parent gene of CircNSD1, NSD1, encodes a protein involved in histone H3 lysine 36 dimethylation (H3K36me) (13, 14). The ChIP assay further revealed a reduced enrichment of H3K36me3, in contrast to H3K36me1 and H3k36me2, within the promoter regions of ACSL4 and SLC39A14 in CircNSD1-overexpressing cells subjected to H/R-induced injured cells. These findings suggest that overexpression of CircNSD1 may attenuate ACSL4 and SLC39A14 DNA H3K36 methylation by H3K36me3 (Figure 6C). IF confirmed CircNSD1 overexpression substantially reduced ACSL4 and SLC39A14 expression in H/R-injured cells and AKI-to-CKD mice (Figure 6, D and E). Together, these findings demonstrate that CircNSD1 overexpression alleviates ferroptosis by downregulating ACSL4 and SLC39A14 in vitro and in vivo.
CircNSD1 encoded a 916aa peptide termed NSD1-916aa. To further explore whether CircNSD1 functioned by encoding a peptide — consistent with emerging evidence of protein-coding potential in certain circRNAs (9) — we performed immunoprecipitation of FLAG-tagged CircNSD1 and luciferase-reporter assays in HK-2 and 293T cells. As predicted by circRNA Db (http://reprod.njmu.edu.cn/circrnadb/), a comprehensive repository for human circRNAs with protein-coding annotations (15), CircNSD1 initiates translation at the ATG start codon and terminates at the TAA stop codon (Figure 7A). This circRNA contains an ORF spanning 2,869 nucleotides, encoding a protein of 916 amino acids protein (916aa). Thus, we named the predicted protein NSD1-916aa, and its sequence is shown in Figure 7B. To further characterize NSD1-916aa, we performed immunofluorescence and MS following the immunoprecipitation and overexpression of FLAG-tagged circNSD1. Immunofluorescence staining of Flag confirmed NSD1-916aa expression in the cytoplasm and nucleus of HK-2 cells (Figure 7C). The MS results of HK-2 cells successfully identified 4 unique sequences of NSD1-916aa, including “QKPLISNSHTDHLMGCTKSAEPGTETSQVNLSDLK,” “EQRLMTAQNLVSYRSPGR,” “KGHIQFEAHKDER,” and “LRDAFSAQMVKNTVNR.” FLAG-tagged CircNSD1 was also transfected into 293T cells, with MS confirming the peptide sequence “QKPLISNSHTDHLMGCTKSAEPGTETSQVNLSDLK.” These results demonstrate that CircNSD1 is translated into the NSD1-916aa protein (Figure 7D and Supplemental Figure 2A).
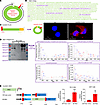 Figure 7
Figure 7CircNSD1 encodes a 916aa peptide. (A) The 2869 nt circNSD1 is potentially translated into a 916aa protein (NSD1-916aa). (B) The sequences indicate the unique amino acids formed by the spanning junction ORF. (C) An illustration of overexpression of FLAG-tagged circNSD1. Immunofluorescence for NSD1-916aa in HK-2 cells. (D) Western blot analysis showing NSD1-916aa in HK-2 cells and the MS results of HK-2 cells revealing 4 unique sequences of NSD1-916aa. (E) Luciferase reporter assay indicating IRES activity of the CircNSD1 UTR exists in HK-2 and 293T cells. Scale bar: 20 μm (white). Data are shown as mean ± SEM, **P < 0.01, ***P < 0.001 compared with the EV group by Student’s t test.
Meanwhile, to validate the internal ribosome entry site (IRES) activity of CircNSD1, a luciferase reporter assay was performed in HK-2 and 293T cells. The promoter drives transcription of firefly luciferase (Fluc) and Renilla luciferase coding regions, with the spacer in bicistronic constructs replaced by either an empty vector or the CircNSD1 untranslated region (UTR) containing the IRES. Results show that intact IRES sequences could trigger Fluc activity, indicating that IRES activity of the CircNSD1 UTR exists (Figure 7E). Collectively, these results confirm that CircNSD1 encoded a 916aa peptide.
-
Discussion
AKI is a clinical syndrome associated with high morbidity and mortality and often progresses to CKD, particularly in cases of medical negligence. Due to the unclear pathogenesis of AKI, targeted treatments remain limited, with hemodialysis being the primary intervention. In the present study, we identified a circRNA, named CircNSD1, which we propose as a potential diagnostic biomarker for AKI-to-CKD transition. CircNSD1 was identified though high-throughput circRNA-Seq of renal cortex tissues, and its downregulation was observed in tissues of AKI-to-CKD mouse model, patients after AKI, and H/R-induced HK-2 cells. Our findings indicate that overexpression of CircNSD1 reduced ferroptosis in vivo and in vitro, thereby slowing the progression of AKI-to-CKD transition. We provide evidence that overexpression of CircNSD1 downregulates ACSL4 and SLC39A14 though the methylation of H3K36, a key mechanism by which CircNSD1 mitigates ferroptosis induced by AKI or H/R. Furthermore, we found that CircNSD1 might encoded a NSD1-916aa peptide, which could have functional implications in its effects. Collectively, these results demonstrate that CircNSD1 is a promising diagnostic and therapeutic target for the early detection of AKI-to-CKD transition (Figure 8).
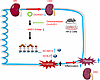 Figure 8
Figure 8Graphical abstract. During the transition from AKI to CKD, CircNSD1 expression decreases and is negatively correlated with inflammation and fibrosis. In vivo and in vitro, overexpression of CircNSD1 mitigates ferroptosis in renal tubular epithelial cells and slows the progression of AKI to CKD. Mechanically, CircNSD1 can encode a NSD1-916aa peptide, which downregulates ACSL4 and SLC39A14 through H3K36 methylation and regulates ferroptosis, inflammation, and fibrosis in AKI to CKD.
In this research, we investigated the role of epithelial-specific CircNSD1 in the AKI-to-CKD transition. circRNAs are highly stable, widely expressed across various cell types, and conserved across species and tissues (16–18). Studies have demonstrated their critical involvement in the pathogenesis, progression, diagnosis, and prognosis of AKI (19, 20). Notably, circRNAs exhibit dynamic, spatiotemporally regulated expression patterns in kidney-related diseases. For instance, Xu et al. reported that circ-AKT3 exacerbates renal I/R injury by sponging miR-144-5p and activating the Wnt/bcatenin pathway and oxidative stress (21). Cheng et al. systematically analyzed the competing endogenous RNA mechanism involving circRNAs and long ncRNAs in a rat model of contrast-induced AKI (CI-AKI), offering insights into the pathogenesis of CI-AKI (22). Although alterations in circRNA expression have been observed in AKI, direct mechanistic links of circRNAs in the AKI-to-CKD transition remain unexplored. This study is the first to our knowledge to reveal that CircNSD1 regulated epithelial injury, with its expression levels being inversely associated with inflammation and fibrosis in AKI-to-CKD patients. These findings suggest that CircNSD1 may serve as a potential diagnostic biomarker for AKI-to-CKD transition.
Growing evidence implicates ferroptosis as a key driver of AKI (23–25). Ferroptosis has been confirmed in mouse models of AKI induced by nephrotoxic drugs and rhabdomyolysis (26–28). Moreover, ferroptotic stress has been identified as a key contributor to the pathogenesis of AKI and its subsequent transition to CKD. Research from Duke University School of Medicine (Durham, North Carolina, USA) has elucidated the molecular mechanisms underlying this transition. Following AKI-induced injury, proximal tubular cells adopt a proinflammatory state that promotes fibrosis. This inflammatory phenotype is transient in cases of mild injury, allowing for a return to the basal state without fibrosis development. However, in instances of severe injury, the inflammatory state becomes chronic, leading to ongoing inflammation and fibrosis. The downregulation of genes involved in glutathione metabolism makes PT cells more susceptible to ferroptotic stress, intensifying the inflammatory response. Genetic induction of high ferroptotic stress after injury exacerbates inflammation and fibrosis, pivotal drivers of AKI-to-CKD transition (6). Our experiments corroborated these findings, demonstrating dynamic changes in ferroptosis markers (e.g., GPX4 and ACSL4) during AKI-to-CKD progression (Supplemental Figure 1, I and J). Collectively, these data underscore ferroptosis as a pathogenic mechanism in this transition. Additionally, untargeted LC-MS/MS analysis of H/R-treated control cells and those overexpressing CircNSD1 revealed a notable enrichment of differential metabolites in the ferroptosis and glutathione metabolism pathways. Furthermore, ChIP assay demonstrated reduced enrichment of H3K36me3 and H3K36me1 at the promoter regions of ACSL4 and SLC39A14 in CircNSD1-overexpressing cells subjected to H/R-induced injury. These results indicate that the overexpression of CircNSD1 may mitigate ferroptosis through the modulation of ACSL4 and SLC39A14 DNA H3K36 methylation patterns during the AKI-to-CKD transition.
Although circRNAs have traditionally been classified as ncRNAs, recent studies indicate that they still have translational functions, and a few circRNAs can be translated into functional proteins under the regulation of methylation modification and participate in a variety of cellular processes (29–31). Notably, protein-coding circRNAs have garnered attention for their involvement in pathogenesis, including muscle atrophy, gastric cancer, AKI, and so on (10, 32–34). For instance, Circ-ZNF609 encoded ZNF609-250aa to induce cell apoptosis and AKI by impairing the autophagy flux via AKT/mTOR-dependent mechanism (9). Despite growing insights into the roles of protein-coding circRNAs in various diseases, their involvement in renal disease progression remain poorly characterized, particularly in the context of the AKI-to-CKD transition. In this study, we identified a protein-coding circRNA, CircNSD1, which initiates translation at the start codon ATG and terminates at the stop codon TAA, encompassing an ORF of 2869 nucleotides. This ORF encodes a protein of 916aa in length, referred to as NSD1-916aa. Our findings suggest that NSD1-916aa, the translational product of CircNSD1, may play a pivotal role in mitigating ferroptosis in the pathogenesis of AKI and CKD.
NSD1, a H3K36 methyltransferase, regulates gene expression through its catalytic activity on histone H3 lysine 36 (H3K36). NSD1 catalyzes the mono- and dimethylation of H3K36, serving as substrates for trimethylation by SETD2. Through interaction with promoter regions and RNA polymerase II (RNAPII), NSD1 modulates transcriptional activity and promotes gene expression by facilitating RNAPII-mediated elongation (35). In this study, overexpression of CircNSD1 reduced H3K36 methylation at ACSL4 and SLC39A14 loci during AKI-to-CKD progression. Overexpression of CircNSD1 exerts effects opposing those of their corresponding linear parent genes (NSD1), potentially due to NSD1-916aa’s interaction with Cullin-associated nedd8-dissociated protein (CAND1) in HK-2 and 293T cells (Supplemental Figure 2, A and B). This interaction between may attenuate the ubiquitination process catalyzed by Cullin 1, a core component of the SCF (SKP1, CUL1/CDC53, F box proteins) E3 ubiquitin ligase complex (36). Stabilization of NSD1-916aa may be essential for CircNSD1’s ability to suppress H3K36 methylation at ACSL4 and SLC39A14 promoters. This epigenetic modulation is critical for mitigating ferroptosis in the context of AKI and subsequently slow the progression to CKD.
In conclusion, our study identifies CircNSD1 as a potential diagnostic biomarker for the AKI-to-CKD transition. CircNSD1 overexpression attenuate ferroptosis in both in vivo and in vitro models, thereby slowing the progression of AKI to CKD. Additionally, the overexpression of CircNSD1 was found to downregulate ACSL4 and SLC39A14 through H3K36 methylation, a critical epigenetic modification that represents a key mechanism in the role of CircNSD1 in ferroptosis induced by AKI or H/R injury. Furthermore, our findings reveal that CircNSD1 encodes a NSD1-916aa peptide, which likely contributes to its functional role in AKI pathophysiology. Collectively, these findings advance our understanding of CircNSD1’s molecular mechanisms in AKI and highlight its therapeutic potential for halting CKD progression.
-
Methods
Sex as a biological variable. Male C57BL/6J mice (purchased from the Experimental Animal Center of Anhui Medical University) were utilized in this study. Our study exclusively examined male mice in this model of AKI to CKD. It is unknown whether the findings are relevant for female mice.
Human samples. Patients with nephropathies who underwent renal biopsies in the Nephrology Department between 2022 and 2024 were screened for inclusion (37, 38). Exclusion criteria for the post-AKI group included: (a) immune-mediated nephropathy; (b) patients diagnosed with AKI who progressed to renal failure or even death; and (c) incomplete clinical data or absence of key laboratory findings.
Inclusion criteria for the post-AKI group were as follows. (a) Diagnosis of AKI per Kidney Disease Improving Global Outcomes (KDIGO) guidelines, meeting one of the following: an increase in serum creatinine > 26.5 μmol/L within 48 hours; or serum creatinine ≥ 1.5× baseline within 7 days; urine output < 0.5 mL/kg/h for at least 6 hours. (b) Post-AKI renal biopsies, stratified into 2 groups based on time from AKI onset: an “early” group (biopsy 2–15 days post-AKI) and a “late” group (biopsy > 15 days after AKI).
The healthy control group was composed of patients with kidney tumors from the urology department, with histologically normal kidney tissues sampled ≥ 5 cm from tumor margins. Inclusion criteria for the control group were as follows: (a) GFR greater than 90 mL/min/1.73 m2; (b) urinary albumin excretion rate (AER) less than 30 mg/d or 20 mg/L, or albumin-to-creatinine ratio (ACR) <30 mg/g; and (c) normal blood pressure, liver function, and complete blood count. Exclusion criteria included: (a) history of metabolic diseases; (b) recent history of nephrotoxic medications; and (c) history of cardiovascular or cerebrovascular diseases. Following screening, 7 post-AKI biopsies (2 early, 5 late) and 7 control biopsies were included.
Animals and animal care. All animal experiments were conducted at Anhui Medical University (Anhui, China) under controlled conditions: temperature 22.5°C ± 0.5°C, humidity 50% ± 5%, a 12-hour light/dark cycle, and ad libitum access to water and a standard murine diet.
To establish the I/R-induced AKI-to-CKD mouse model, bilateral renal pedicles were clamped for 30 minutes using nontraumatic microaneurysm clamps (Roboz) to induce ischemia, followed by reperfusion. Kidneys were repositioned, and incisions were closed with absorbable sutures and wound clips. Sham surgeries were performed on control mice (n = 6–8). GFR was determined by measuring the transcutaneous elimination of FITC-sinistrin as described previously (39). Briefly, under brief isoflurane anesthesia, a fluorescence detector (MediBeacon) was placed on a depilated dorsal region, and FITC-sinistrin (50 mg/kg) was injected i.v. via the tail vein. Following anesthesia with sodium pentobarbital (50 mg/kg, i.p.), kidney and blood samples were collected for further experiments.
Eight-week-old male C57BL/6J mice were randomly divided into 2 groups: (a) mice injected with the HBAAV2/9-Null (EV, n = 6–8) and (b) mice injected with the HBAAV2/9-mmu_circ_0000465 (CircNsd1-OE, n = 6–8). A recombinant adeno-associated virus containing the mouse CircNsd1-OE gene was purchased from Hanheng Company (Hanheng Biotechnology Co.). The 150 UL of 1.7 × 1012 vg/mL adeno-associated virus 2/9 carrying the CircNsd1-OE or EV was injected by tail vein in mice (Supplemental Table 2). Two weeks after AAV2/9 administration, renal ischemia was induced by clamping the renal pedicle for 30 minutes. After removing the clamps to allow reperfusion, the kidneys were repositioned into the peritoneal cavity. At 12 weeks of age, mice were anesthetized with sodium pentobarbital (50 mg/kg, i.p.), and kidney tissues and blood samples were collected for analysis.
Reagents and materials. The following antibodies were used: anti–BACTIN antibody (Proteintech, 66009-1-Ig), anti-ACSL4 antibody (Santa Cruz Biotechnology, A0223), anti-GPX4 antibody (HUABIO, ET1706-45), anti-CD28K antibody (Proteintech, 66394-1-Ig), anti-AQP3 antibody (Santa Cruz Biotechnology, sc-518001), anti-SLC39A14 antibody (Proteintech, 26540-1-AP), anti–mouse Flag (Proteintech, 10024351), anti-H3K36me1 (Abcam, ab9048), anti-H3K36me2 (Abcam, ab9049), anti-H3K36me3 (Abcam, ab9050), anti–fluorescein labeled lotus tetragonolobus lectin (LTL) antibody (Vector Laboratories, FL-1321), anti-F4/80 antibody (Cell Signaling Technology, D2S9R), anti-IgG antibody (Cell Signaling Technology, 38191s-9), anti-ASMA (ZSbio, Kit-0006), and anti-CD68 antibody (Abcam, ab955). Masson’s trichrome staining kits were procured from Zhuhai Besso Biotechnology Institute. The PAS kits were obtained from Solarbio.
Cell culture. Human kidney tubular epithelial cell lines (HK-2 cells) were provided by Huiyao Lan (the Chinese University of Hong Kong, Hong Kong, China). The Cells were cultured in DMEM/F12 medium supplemented with 5% FBS (Gibco) at 37°C in a humidified atmosphere with 5% CO2. Following serum starvation, cells were transferred to an anoxic chamber (94% N2, 5% CO2, 1% O2) for 24 hours, before being returned to normoxic conditions. At 48 hours after reoxygenation, cells were harvested and analyzed for ferroptosis markers, oxidative stress, and inflammatory responses using real-time PCR and related assays.
RNA extraction and real-time PCR. Total RNA was extracted from kidney homogenates and HK-2 cells using TRIzol reagent (Takara) following the manufacturer’s instructions. RNA concentrations were determined using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from total RNA using RealMasterMix (Yeasen) with nuclease-free water. Real-time PCR was performed using the Hieff UNICON qPCR SYBR Mix (Yeasen), as previously described (40, 41). Amplification and detection of cDNA were carried out using a Thermal Cycler system (CFX-6, Bio-Rad). β-Actin served as an internal reference gene for normalization, and the data are expressed as the mean ± SEM. Primer sequences used in this study are detailed in Supplemental Table 1. The relative quantification of gene expression was calculated using the 2–ΔΔCt method.
Dual-luciferase reporter assay. Predicted sequences for the IRES were cloned into the Luc-IRES-Report vector (Hanbio). The sequence used was: 5′-TAAGAAAAAGTCTACGCCACTGAAGTATGAAGTTGGAGATCTCATCTGGGCAAAATTCAAGAGACGCCCATGGTGGCCCTGCAGGATTTGTTCTGATCCGTTGATTAACACACATTCAAAAATG-3′. Luciferase activity was measured using the Dual-Glo Luciferase Assay Kit (Promega) to quantify firefly and Renilla luciferase activities, following the manufacturer’s protocol (42, 43).
Immunoprecipitation. HK-2 Cells were harvested and washed thrice with ice-cold PBS as previously described (44). The cells were lysed in IP buffer (Beyotime Biotechnology) supplemented with a complete protease inhibitor cocktail (PIC) (Yeasen) on ice for 30 minutes. Lysates were collected, and immunoprecipitation was performed using anti-Flag Nanoab Magarose beads (NuoyiBio) by incubating with protein samples at 4°C for 12 hours. The resultant immunoprecipitated proteins were analyzed using gel electrophoresis and LC-MS/MS for identification and quantification, as reported previously (45).
LC-MS/MS. Immunoprecipitated proteins extracted from SDS-PAGE gels were subjected to enzymatic digestion, following established protocols. Gel pieces were washed with 50% acetonitrile (ACN) for 30 minutes, followed by a second wash with 100% ACN for an additional 30 minutes. After reduction and alkylation, the proteins were digested with trypsin at 37°C for 12 hours. The supernatant containing the digested peptides was collected and processed for desalination using an HPLC buffer system and analyzed by LC-MS/MS on Easy-nLC1200 chromatograph (Thermo Fisher Scientific) coupled to a Q Exactive plus mass spectrometer (Thermo Fisher Scientific). Data analysis was performed using Proteome Discoverer 2.4, incorporating the predicted sequence of NSD1-916aa and searching against the UniProt-reviewed Homo sapiens database. The experiments were conducted by OE Biotechnology Company, with data deposited in the National Genomics Data Center.
Immunofluorescence. Cells were fixed with 4% PFA for 5 minutes, rinsed with PBS, and blocked with 10% donkey serum for 60 minutes at 37°C. Subsequently, the cells were incubated with primary antibodies against α-SMA/Acsl4/SLC39A14 overnight at 4°C. After primary antibody incubation, the cells were treated with a FITC-conjugated secondary antibody (Bioss). Nuclei were counterstained with DAPI. After a final PBS wash, the sections were scanned using an automatic digital slide scanner (Pannoramic Scan).
Lentivirus package and transfection of CircNSD1 overexpression. The circNSD1-FLAG or control sequence was cloned into the pHBLV-CMV-circ-EF1-puro vector (Supplemental Table 3). This recombinant vector, along with packaging plasmids psPAX2 and pMD2.G, was cotransfected into HEK293T cells at a ratio of 4:3:1. After 48 hours, lentivirus particles were harvested from the culture supernatant and filtered. HK-2 cells or 293T cells were infected with the lentivirus, and stable cell lines were established by selection with puromycin (10 μg/mL) at 48 hours after infection.
ChIP-qPCR. To confirm that H3k36me1, H3k36me2, and H3k36me3 protein directly binds to the promoter of ACSL4 and SlC39A14, ChIP assays were performed using the Enzymatic ChIP kit (Cell Signaling Technology). Cells were initially fixed with a crosslinking agent to stabilize protein-DNA interactions. Following crosslinking, cells were washed twice with cold PBS to remove excess crosslinking agent; they were then rinsed once with 2 mL of cold PBS supplemented with 10 μL of PIC to prevent proteolysis. Cells were subsequently lysed, and nuclei were isolated by centrifugation at 16,000g. The crosslinked chromatin was then fragmented by ultrasonication into DNA fragments ranging from 150 to 900 bp. ChIP was conducted by incubating samples with antibodies specific to anti-p65 or a nonspecific IgG (as a negative control) overnight at 4°C. DNA samples were purified using magnetic beads and subjected to ChIP–quantitative PCR (ChIP-qPCR) using the following primers: ACSL4, forward (F), 5′-GCGGGGATCTTAACGCAGTA-3′ and reverse (R), 5′-GTTCCCCTCGGGTCATCTTC-3′; SlC39A14, F, 5′-CCTCGGAACTTCTTCACCCC-3′ and R, 5′-TCCCACAATGATCAGACGCC-3′; LPCAT3, F, 5′-AGAGGAAATGCTAGGCCCGT-3′ and R, 5′-GTGTGCGCGAATCTGTGTTC-3′; and SAT1, F, 5′-CCAGTAGGGTTTCCGCCAAG-3′ and R, 5′-TGAACCCGGAGGACAAAAGT-3′.
Untargeted LC/MS lipidomics analysis. Lipids were extracted from H/R-treated control and circNSD1-overexpressing cells using a standardized protocol. Quality control (QC) volume was prepared by pooling equal volumes of extracts from all samples, ensuring that the volume of each QC sample was equivalent to that of the individual samples. Subsequently, the samples were subjected to separation using a Nexera UPLC system (Shimadzu) and analyzed via a Q Exactive mass spectrometer (Thermo Fisher Scientific). The LC/MS detection was performed at Lumingbio Company. A supervised multiple regression OPLS-DA model was applied to identify differential lipid metabolites. Permutation testing with 200 iterations was conducted to mitigate overfitting risks. All experiments were conducted by OE Biotechnology Company and data are available at the National Genomics Data Center (https://ngdc.cncb.ac.cn/omix/release/OMIX007361).
Kidney histology. Kidney tissue sections were prepared from paraffin-embedded samples and subjected to IHC staining. Antigen retrieval was performed using a microwave-based method. Sections were incubated with rabbit-derived primary antibodies against ASMA, F4/80, or CD68 for 24 hours at 4°C, followed by incubation with appropriate secondary antibodies for 30 minutes at 37°C. Staining was visualized using 3,3’-diaminobenzidine (DAB) as the chromogen. Slides were scanned using a Pannoramic Scan digital slide scanner (3D Histech).
Evaluation of renal damage. Human and mouse kidney tissues were fixed in 4% paraformaldehyde (PFA), dehydrated, and embedded in paraffin. Tissue sections (5 μm thick) were stained with H&E to assess morphology, PAS to evaluate tissue damage, and Masson’s trichrome to quantify fibrosis, following the manufacturer’s protocol (46).
Renal damage was assessed using PAS- and H&E-stained kidney specimens. Parameters evaluated included tubular detachment, tubular dilation, protein casts, and brush border damage. Six random fields per section were analyzed, with results expressed as the percentage of affected area per whole section. The extent of tubular detachment, proximal tubular dilation, and brush border damage was scored on a scale from 0 to 10, with the following categorization: 0–1 indicating none (0%), 1–2 indicating less than 11%, 2–4 indicating 11%–25%, 4–6 indicating 26%–45%, 6–8 indicating 46%–75%, and 8–10 indicating greater than 75% (47).
Detection of circNSD1 levels in serum. To measure circNSD1 levels in serum, samples from patients and controls were processed as follows. Serum was centrifuged at 2,000g for 30 minutes at 4°C, and the supernatant was collected. The supernatant was further centrifuged at 12,000g at 4°C for 45 minutes, filtered through a 0.22 μm filter (MilliporeSigma), and ultracentrifuged at 110,000g for 70 minutes at 4°C (Beckman Ultracentrifuge). The pellet was washed with PBS, recentrifuged at 110,000g for 70 minutes, and resuspended in 500 μL of TRIzol reagent. Total RNA was extracted, and circNSD1 levels were quantified by real-time PCR according to standard protocols.
Statistics. Data are presented as the mean ± SEM (38). Statistical significance was determined using 1-way ANOVA, followed by Tukey’s post hoc test for multiple comparisons. Additionally, 2-tailed Student’s t test were also performed to compare the means of 2 groups when necessary, as performed with GraphPad Prism version 5.0 software (GraphPad Software).
Study approval. The clinical studies were performed in accordance with the principles of the Declaration of Helsinki after obtaining informed consent from the patients. It was approved by the Research Ethics Committee of the First Affiliated Hospital of Anhui Medical University (no. 2023187). All animal experiments were approved by the Anhui Medical University Ethical Committee on Animal Experiments (no. LLSC20211279).
Data availability. The datasets used or analyzed in the study are available from the Supporting Data Values file. The high-throughput circRNA-Seq data in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139107). The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier OMIX007182, OMIX007357, and OMIX007361 (https://ngdc.cncb.ac.cn/omix/release/OMIX007182; https://ngdc.cncb.ac.cn/omix/release/OMIX007357; https://ngdc.cncb.ac.cn/omix/release/OMIX007361). All original data are accessible through the corresponding author upon reasonable request.
-
Author contributions
LG, JZ, CC and SZ contributed methodology, investigation, review and editing, writing of the original draft, and formal analysis. XW, GT, SW, Y Wang, and XL contributed review and editing as well as methodology. LJ and Y Wu contributed conceptualization, supervision, project administration, and funding acquisition. All authors reviewed the manuscript.
-
Acknowledgments
We acknowledge the use of an online analyzer for kidney single-cell datasets, named Kidney Interactive Transcriptomics (KIT). This study was supported by the National Natural Science Foundation (no. 82300820) and the China Postdoctoral Science Foundation (nos. 2023T160001 and 2022M710179) though funding awarded to LG; this study was also supported by the National Natural Science Foundation (no. 82200806), 2022 Anhui Provincial Department of Education University research project (no. 2022AH051180), and 2022 Anhui Province translational medicine project cultivation project (no. 2022zhyx-C31) though funding awarded to LJ. Additionally, it received support from the Cultivation Fund of the First Affiliated Hospital of Anhui Medical University, with grant number 2024YKJ02, awarded to JZ.
Address correspondence to: Yonggui Wu or Ling Jiang, Department of Nephropathy, The First Affiliated Hospital of Anhui Medical University, No. 218, JiXi Road, Hefei, Anhui, 230022, China. Phone: 86.0551.6292.2111; Email: wuyonggui@medmail.com.cn (YW); Email: jiangling0022@163.com (LJ).
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Authorship note: LG, JZ, CC, and SZ contributed equally to this work.
Copyright: © 2025, Gao et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2025;10(16):e189130.https://doi.org/10.1172/jci.insight.189130.
-
References
- Zarbock A, et al. Epidemiology of surgery associated acute kidney injury (EPIS-AKI): a prospective international observational multi-center clinical study. Intensive Care Med. 2023;49(12):1441–1455.
- Gao L, et al. Potential targeted therapy and diagnosis based on novel insight into growth factors, receptors, and downstream effectors in acute kidney injury and acute kidney injury-chronic kidney disease progression. Signal Transduct Target Ther. 2020;5(1):9.
- Emily JS, et al. Long-term risk of adverse outcomes after acute kidney injury: a systematic review and meta-analysis of cohort studies using consensus definitions of exposure. Kidney Int. 2018;95(1):160–172.View this article via: PubMed Google Scholar
- Shuiqin G, et al. REST contributes to AKI-to-CKD transition through inducing ferroptosis in renal tubular epithelial cells. JCI Insight. 2023;8(11):e166001.
- Manjeri AV, et al. Failed tubule recovery, AKI-CKD transition, and kidney disease progression. J Am Soc Nephrol. 2015;26(8):1765–1776.
- Shintaro I, et al. Ferroptotic stress promotes the accumulation of pro-inflammatory proximal tubular cells in maladaptive renal repair. Elife. 2021;10:e68603.
- Yuhei K, et al. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc Natl Acad Sci U S A. 2020;117(27):15874–15883.
- Pietro EC, et al. Transcriptional trajectories of human kidney injury progression. JCI Insight. 2018;3(22):e123151.
- Xin O, et al. A protein encoded by circular ZNF609 RNA induces acute kidney injury by activating the AKT/mTOR-autophagy pathway. Mol Ther. 2022;31(6):1722–1738.View this article via: PubMed Google Scholar
- Haohan L, et al. Circular MTHFD2L RNA-encoded CM-248aa inhibits gastric cancer progression by targeting the SET-PP2A interaction. Mol Ther. 2023;31(6):1739–1755.
- Jian Z, et al. Circular RNA encoded MET variant promotes glioblastoma tumorigenesis. Nat Commun. 2023;14(1):4467.
- Guo R, et al. The road from AKI to CKD: molecular mechanisms and therapeutic targets of ferroptosis. Cell Death Dis. 2023;14(7):426.
- Shao R, et al. H3K36 methyltransferase NSD1 regulates chondrocyte differentiation for skeletal development and fracture repair. Bone Res. 2021;9(1):30.
- Li Y, et al. Histone methylation antagonism drives tumor immune evasion in squamous cell carcinomas. Mol Cell. 2022;82(20):3901–3918.
- Chen X, et al. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985.
- Jia Y, et al. m6A-modified circCacna1c regulates necroptosis and ischemic myocardial injury by inhibiting Hnrnpf entry into the nucleus. Cell Mol Biol Lett. 2024;29(1):140.
- Conn V, et al. Circular RNA in cancer. Nat Rev Cancer. 2024;24(9):597–613.
- Misir S, et al. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29(3):481–491.
- Li Z, et al. Potential therapeutic applications of circular RNA in acute kidney injury. Biomed Pharmacother. 2024;174:116502.
- So B, et al. Circular RNAs in acute kidney injury: roles in pathophysiology and implications for clinical management. Int J Mol Sci. 2022;23(15):8509.
- Xu Y, et al. circ-AKT3 aggravates renal ischaemia-reperfusion injury via regulating miR-144-5p /Wnt/β-catenin pathway and oxidative stress. J Cell Mol Med. 2022;26(6):1766–1775.
- Cheng W, et al. Non-coding RNA-associated ceRNA networks in a new contrast-induced acute kidney injury rat model. Mol Ther Nucleic Acids. 2019;17:102–112.
- Gao G, et al. Carbon dot nanozymes with ferrous ion-chelating and antioxidative activity inhibiting ferroptosis to alleviate renal ischemia-reperfusion injury. Small. 2025;21(16):e2407372. View this article via: PubMed Google Scholar
- Ni L, et al. Targeting ferroptosis in acute kidney injury. Cell Death Dis. 2022;13(2):182.
- Hu Z, et al. Emerging role of ferroptosis in acute kidney injury. Oxid Med Cell Longev. 2019;2019:8010614. View this article via: PubMed Google Scholar
- Martin-Sanchez D, et al. Ferroptosis, but not necroptosis, is important in nephrotoxic folic acid-induced AKI. J Am Soc Nephrol. 2017;28(1):218–229.
- Guerrero-Hue M, et al. Curcumin reduces renal damage associated with rhabdomyolysis by decreasing ferroptosis-mediated cell death. FASEB J. 2019;33(8):8961–8975.
- Mishima E, et al. Drugs repurposed as antiferroptosis agents suppress organ damage, including AKI, by functioning as lipid peroxyl radical scavengers. J Am Soc Nephrol. 2020;31(2):280–296.
- Liu CX, et al. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185(13):2390.
- Margvelani G, et al. Translation of circular RNAs. Nucleic Acids Res. 2025;53(1):gkae1167.
- Lan T, et al. The protein circPETH-147aa regulates metabolic reprogramming in hepatocellular carcinoma cells to remodel immunosuppressive microenvironment. Nat Commun. 2025;16(1):333.
- Chen R, et al. CircTmeff1 promotes muscle atrophy by interacting with TDP-43 and encoding a novel TMEFF1-339aa protein. Adv Sci (Weinh). 2023;10(17):e2206732. View this article via: PubMed Google Scholar
- Legnini I, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.
- Qiao L, et al. Inhibitory effects of circR-127aa on gastric cancer progression and tumor growth. Cell Signal. 2025;125:111520.
- Krossa I, et al. Lysine methyltransferase NSD1 and cancers: any role in melanoma? Cancers (Basel). 2022;14(19):4865.
- Huang X, et al. Cullin-associated and neddylation-dissociated protein 1 (CAND1) alleviates NAFLD by reducing ubiquitinated degradation of ACAA2. Nat Commun. 2023;14(1):4620.
- Ana SH, et al. Infectious consequences of the AKI-to-CKD transition. Clin Kidney J. 2022;15(12):2237–2244.
- Letizia DC, et al. Tubular cell polyploidy protects from lethal acute kidney injury but promotes consequent chronic kidney disease. Nat Commun. 2022;13(1):5805.
- Li G, et al. STING/ACSL4 axis-dependent ferroptosis and inflammation promote hypertension-associated chronic kidney disease. Mol Ther. 2023;31(10):3084–3103.
- Xiaomeng F, et al. SIRT3 deficiency sensitizes angiotensin-II-induced renal fibrosis. Cells. 2020;9(11):2510.
- Jiang L, et al. METTL3-mediated m6A modification of TIMP2 mRNA promotes podocyte injury in diabetic nephropathy. Mol Ther. 2022;30(4):1721–1740.
- Xueqi L, et al. Circ-0000953 deficiency exacerbates podocyte injury and autophagy disorder by targeting Mir665-3p-Atg4b in diabetic nephropathy. Autophagy. 2023;20(5):1072–1097.View this article via: PubMed Google Scholar
- Ling J, et al. hsa-miR-500a-3P alleviates kidney injury by targeting MLKL-mediated necroptosis in renal epithelial cells. FASEB J. 2018;33(3):3523–3535.View this article via: PubMed Google Scholar
- Li G, et al. THBS1/CD47 modulates the interaction of γ-catenin with E-cadherin and participates in epithelial-mesenchymal transformation in lipid nephrotoxicity. Front Cell Dev Biol. 2021;8:601521.
- Xue-Qi L, et al. Wogonin protects glomerular podocytes by targeting Bcl-2-mediated autophagy and apoptosis in diabetic kidney disease. Acta Pharmacol Sin. 2021;43(1):96–110.View this article via: PubMed Google Scholar
- Li G, et al. Restoration of E-cadherin by PPBICA protects against cisplatin-induced acute kidney injury by attenuating inflammation and programmed cell death. Lab Invest. 2018;98(7):911–923.
- Wolak T, et al. Osteopontin modulates angiotensin II-induced inflammation, oxidative stress, and fibrosis of the kidney. Kidney Int. 2009;76(1):32–43.
-
Version history
- Version 1 (July 15, 2025): In-Press Preview
- Version 2 (August 22, 2025): Electronic publication












