Clinical Research and Public HealthClinical ResearchOncology
Open Access |  10.1172/jci.insight.194823
10.1172/jci.insight.194823
First-in-child phase I trial of p-STAT3 inhibitor WP1066 in pediatric brain tumor patients
Robert C. Castellino,1,2 Hope Mumme,3 Andrea Franson,4 Bing Yu,1,2 Hope Robinson,1,2 Kavita Dhodapkar,1,2 Dolly Aguilera,1,2 Matthew Schniederjan,2,5 Rohali Keesari,2 Zhulin He,2 Manoj Bhasin,1,2,3 Waldemar Priebe,6 Amy B. Heimberger,7 and Tobey J. MacDonald1,2
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Castellino, R. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Mumme, H. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Franson, A. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Yu, B. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Robinson, H. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Dhodapkar, K. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Aguilera, D. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Schniederjan, M. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Keesari, R. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by He, Z. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Bhasin, M. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by Priebe, W. in: PubMed | Google Scholar
1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by
Heimberger, A.
in:
PubMed
|
Google Scholar
|

1Aflac Cancer and Blood Disorders Center, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
2Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
3Department of Biomedical Informatics, Emory University, Atlanta, Georgia, USA.
4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, Michigan, USA.
5Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia, USA.
6Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
7Department of Neurological Surgery, Malnati Brain Tumor Institute of the Robert H. Lurie Comprehensive Cancer Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois, USA.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
Find articles by
MacDonald, T.
in:
PubMed
|
Google Scholar
|

Published December 11, 2025 - More info
JCI Insight. 2026;11(3):e194823. https://doi.org/10.1172/jci.insight.194823.
© 2026 Castellino et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: April 24, 2025; Accepted: December 9, 2025
-
Abstract
BACKGROUND. WP1066 is an orally bioavailable, small-molecule inhibitor of activated phosphorylated STAT3 (p-STAT3) that has demonstrated preclinical efficacy in pediatric brain tumor models.
METHODS. In a first-in-child, single-center, single-arm 3+3 design phase I clinical trial, 10 patients were treated with WP1066 twice daily, Monday-Wednesday-Friday, for 14 days of each 28-day cycle to determine the maximum tolerated dose/maximum feasible dose of WP1066. Compassionate-use treatment with WP1066 in 3 pediatric patients with H3.3G34R/V-mutant high-grade glioma (HGG) is also described.
RESULTS. There was no significant toxicity, and the maximum feasible dose (MFD) was determined to be 8 mg/kg. Treatment-related adverse events were grade 1–2 (diarrhea and nausea most common); there were no dose-limiting toxicities. Median progression-free and overall survival was 1.8 months and 4.9 months, respectively. One partial response was observed in a patient with pontine glioma. Among the H3.3G34R/V-mutant HGG patients not on study, WP1066 was administered after upfront radiation to one patient for 17 months. At all dose levels tested, WP1066 suppressed p-STAT3 expression by peripheral blood mononuclear cells (PBMCs). Single-cell RNA sequencing analysis of PBMCs demonstrated increased CD4+ and CD8+ T cells, proinflammatory TNFA signaling, differentiation activity in myeloid cells, and downregulation of Tregs after WP1066 treatment, consistent with systemically inhibited STAT3 activity.
CONCLUSION. WP1066 is safe, has minimal toxicity, and induces antitumor immune responses in pediatric brain tumor patients. Phase II investigation of WP1066 at the MFD in this patient population is warranted.
TRIAL REGISTRATION. ClinicalTrials.gov NCT04334863.
FUNDING. CURE Childhood Cancer and Peach Bowl Inc.
-
Introduction
Pediatric high-grade glioma (pHGG), which includes diffuse midline glioma (DMG) and diffuse intrinsic pontine glioma (DIPG), accounts for the majority of deaths related to pediatric brain tumors (1). DMG and DIPG have a dismal median overall survival (OS) of 9–11 months from diagnosis (2). Focal radiation therapy (RT) has proven benefit, albeit transient, when administered at diagnosis or at recurrence for pHGG, but 5-year OS remains lower than 20% (3). Relapsed medulloblastoma (MB) and ependymoma (EPN) are also typically fatal and have no accepted standard therapy at recurrence (4, 5). Thus, each of the 3 most common pediatric malignant brain tumor entities, pHGG, MB and EPN, is in urgent need of novel therapies.
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor that plays critical roles in immune regulation and oncogenesis and is overexpressed in central nervous system (CNS) tumors, including pHGG, MB, and EPN (5–9). STAT3 promotes tumorigenesis by preventing apoptosis and enhancing proliferation, angiogenesis, invasion, and metastasis (10). Growth factors and cytokines activate STAT3 by phosphorylating the tyrosine 705 residue in the STAT3 transactivation domain (p-STAT3), resulting in STAT3-induced expression of target genes (11). Gliomas have high levels of p-STAT3 relative to normal brain tissue, and p-STAT3 expression correlates with tumor grade and decreased survival (12–15).
In H3K27M- and H3.3G34R/V-mutant pHGGs, p-STAT3 is selectively upregulated, and STAT3 signaling promotes survival of H3.3G34R/V-mutant glioma cells (16, 17). Likewise, STAT3 knockdown or small-molecule inhibition of STAT3 reduces pHGG stem cell viability and restores expression of the polycomb repressive mark H3K27me3 (18), while STAT3 downregulation in patient-derived DIPG cells decreases cell viability, migration, and invasion (9). Furthermore, radiation has been shown to increase the expression and activation of STAT3 in gliomas (19–21), and combining radiation with STAT3 inhibition significantly increased monotherapy efficacy in HGG models (22). MBs also have high levels of STAT3 (23), and in sonic hedgehog–type (SHH) MB, we have shown that SHH ligand induces expression and activation of STAT3, which is critical to maintaining SHH signaling, promoting cell viability in vitro, and tumor formation in vivo (24).
STAT3 is also a key regulator of immunosuppression and is therefore considered a potential target for cancer immunotherapy (25). STAT3 signaling is induced in immune cells within the tumor microenvironment (TME) (15, 26), which downregulates antitumor immune responses. Immune cells that return to circulation can thus serve as surrogate markers for the TME, as evidenced by significantly increased p-STAT3 expression in peripheral blood mononuclear cells (PBMCs) in patients with glioblastoma multiforme (GBM) (8). p-STAT3 suppresses activation of macrophages (MPs), inhibits natural killer (NK) cell and neutrophil cellular cytotoxicity, and reduces expression of major histocompatibility complex II, CD80, CD86, and interleukin-12 in dendritic cells, rendering them unable to stimulate T cells or to generate effective antitumor immunity (27, 28). STAT3 activation enhances the suppressive activities of human Tregs by upregulating FOXP3 expression (29), and tumor-associated MPs and microglia become polarized via the STAT3 pathway toward tumor-supportive phenotypes (M2) that contribute to angiogenesis and tumor invasion (30).
WP1066 (Moleculin Inc.), a small-molecule congener of caffeic acid benzyl ester able to cross the blood-brain barrier (BBB), is a potent inhibitor of p-STAT3 (31, 32). Unlike most other STAT3 inhibitors, which suppress specific kinase activity upstream of STAT3, WP1066 suppresses p-STAT3 independently of its method of activation in vitro and in vivo. This results in HGG cell apoptosis by downregulating the anti-apoptotic proteins MCL1, BCLXL, and MYC, while activating pro-apoptotic BAX (33). WP1066 can also inhibit vascular endothelial growth factor (VEGF) production and tumor angiogenesis (34, 35). Multiple studies have demonstrated the therapeutic efficacy of WP1066 in various malignancies, including head and neck (36), pancreatic (37), bladder (38), lymphoma (39), gastric (40), CNS melanoma (41), and glioma (42, 43). In genetically engineered murine (GEM) HGG models treated with WP1066, there was a 55.5% increase in median survival, with an associated inhibition of intratumoral p-STAT3 and MPs (42). We have shown that the mechanism of immunologic antitumor cytotoxicity by WP1066 is mediated by a combination of inhibition of Tregs, upregulation of costimulatory CD80 and CD86 molecules on human microglia, induction of proinflammatory cytokine secretion essential for T effector responses, inhibition of immune suppressive cytokines, and the inducement of impaired CD8+ effector T cells to become activated and proliferate (22, 31, 41–44). In both orthotopic and GEM models of H3K27M-mutant DIPG and H3.3G34R/V-mutant HGG, WP1066 treatment significantly extended overall survival (16, 17).
Our group previously reported the results of a first-in-human phase I clinical trial of WP1066 in 8 adult patients with recurrent GBM (NCT01904123) (45). WP1066 was found to be well tolerated with a maximum feasible dose (MFD) of 8 mg/kg. Here, we report the results from our first-in-child phase I clinical trial using an identical dosing schedule of WP1066 to treat pediatric patients with recurrent or progressive high-grade brain tumors refractory to standard treatment.
-
Results
Study enrollment and patient characteristics. The patient flow through the study is shown in the CONSORT diagram (Figure 1). Of 13 patients who gave consent and were screened, 10 patients were eligible and enrolled in the AflacST1901 study between May 1, 2020, and October 26, 2022 (Table 1). Two patients were deemed ineligible because of tumor size or neurologic status at the time of consent. One declined to proceed with trial participation. All patients enrolled in the study had been previously treated with at least standard-of-care therapy, including multi-agent, systemic chemotherapy, and/or radiation therapy. Only 2 patients were receiving corticosteroids at the time of their study treatment (AflacST1901-01 and -010, DIPG and thalamic DMG, respectively). Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.194823DS1) shows patient demographics and tumor characteristics for 3 young adult patients with supratentorial H3.3G34R/V-mutant HGG who were treated off-study with compassionate-use WP1066 at the University of Michigan C.S. Mott Children’s Hospital.
Adverse events and maximum tolerated dose/maximum feasible dose. There were no dose-limiting toxicities (Table 2) and no treatment-related deaths. The most common treatment-related adverse events were grade 1–2 nausea, vomiting, and diarrhea. The most common treatment-related adverse laboratory events were grade 1–2 anemia, white cell count suppression, and elevation of alanine aminotransferase (Table 3). All patients on this pediatric trial reported displeasure with the taste of the liquid formulation of WP1066, and in keeping with the adult phase I trial of WP1066, the maximum tolerated dose/maximum feasible dose (MFD) was determined to be 8 mg/kg. This was also the optimal dose determined for the phase II adult study of WP1066 with upfront radiation. Therefore, the last planned dose level (16 mg/kg) was not investigated in this pediatric trial.
Patient outcomes. The median progression-free survival was 1.8 months, and the median OS was 4.9 months (Figure 2). Of the 10 patients on study with radiographic follow-up, all had eventual radiographic progressive disease and died of disease. However, one patient with progressive DIPG (AflacST1901-01), who had completed a 2-week course of palliative focal re-irradiation more than 4 weeks before enrollment, and had stable tumor size on the baseline pre-study MRI obtained 12 weeks after irradiation and 2 days before starting therapy, subsequently had a 33.8% reduction in maximal tumor volume by MRI by Response Assessment in Neuro-Oncology (RANO) criteria after 2 cycles of treatment with WP1066 (Figure 3). This patient was able to be successfully weaned off high-dose corticosteroids and improved clinically from being wheelchair bound to walking with assistance. A brain MRI after cycle 3 of WP1066 showed a 61.8% increase in tumor size from baseline, but no increase in intratumoral blood perfusion, which was interpreted to be most consistent with treatment-related changes off steroids rather than tumor progression. Per the study protocol, the subject was taken off study treatment at that time because of the increased tumor dimensions; however, the patient continued to exhibit clinical improvement for another 6 months without additional therapy.
 Figure 2
Figure 2Clinical outcomes of pediatric patients treated with WP1066. (A and B) Kaplan-Meier analysis is shown for progression-free survival of patients with clinical and radiographic progression based on MRI scans (A) and overall survival (OS) of pediatric patients with progressive high-grade brain tumors who were treated on study AflacST1901 with WP1066 (B). Shaded area denotes 95% CI. (C) Swimmer plot of treatment duration and events for patients on study.
 Figure 3
Figure 3Radiographic tumor response to WP1066 treatment. Serial brain MRIs from a pediatric patient with progressive DIPG (AflacST1901-01) who had completed a 2-week course of palliative focal re-irradiation to the tumor 12 weeks before starting study treatment show 33.8% reduction in tumor volume after 2 cycles of WP1066 treatment compared with baseline MRI obtained 2 days before the start of study treatment (15.48 cm3 to 10.25 cm3). The patient was successfully tapered off high-dose corticosteroids and improved clinically, from being wheelchair bound to walking with assistance. Subsequent brain MRI performed after 3 cycles of WP1066 while off steroids for 4 weeks (pre–cycle 4) showed a 61.8% increase in tumor volume from baseline (25.05 cm3), but no increase in intratumoral blood perfusion, suggesting that the increase in tumor volume was due to treatment-related changes. Because of the increase in tumor volume, the patient was taken off protocol therapy per the study guidelines; however, the patient continued to exhibit clinical improvement followed by stability for an additional 6 months.
For the 3 young adults with hemispheric H3.3G34R/V-mutant HGG who were treated off-study with WP1066 (8 mg/kg) on a compassionate-use basis, the median OS was 4.5 months (Supplemental Figure 1). One patient who was treated with WP1066 after completion of upfront standard radiation and concurrent temozolomide continued adjuvant WP1066 for 17 months before tumor progression.
STAT3 inhibition and immunologic response to WP1066. Flow cytometric analysis of PBMCs for phosphorylated STAT3 (p-STAT3), which indicates STAT3 activation, demonstrated a significant reduction in p-STAT3 within 24 hours of WP1066 treatment in all patients analyzed (n = 5, AflacST1901-04, -05, -06, -07, and -08), confirming on-target activity of WP1066 in these pediatric patients (Figure 4). Analyses of p-STAT3 in PBMCs by day 8 (D8) of therapy showed that the p-STAT3 signal had reverted to baseline at cycle 1, day 1 (C1D1), hour 0, and the subsequent decreases in p-STAT3 following treatment for C1D8 at 4 hours and 24 hours were virtually identical to that at C1D1 at these same time points (data not shown), indicating that there is not a prolonged or cumulative effect on p-STAT3 with each successive dose of WP1066. Analysis by scRNA-seq of 17 PBMC samples from the 5 corresponding study patients with PBMC p-STAT3 flow cytometric analysis, plus 1 additional subject (AflacST1901-010), demonstrated an overall increase in T and B cells before initiation of treatment to the start of cycle 2 on treatment (Figure 5, Supplemental Table 2, and Supplemental Figure 2). Of the patients analyzed, only subject AflacST1901-010 had received concurrent corticosteroids during protocol therapy, and the dosage of steroids had been stable since 1 week before the start of WP1066 therapy, indicating that the changes observed in immune cells with treatment were not related to concomitant changes in the dosage of steroids administered. scRNA-seq further showed an increase of gene signatures consistent with an increase in naive CD4+ (Figure 6A) and CD8+ T cells (Figure 6B) and a decrease in the Treg signature within the CD4+ T cell compartment (Figure 6C) consistent with reduced STAT3 activation. Meanwhile, the gene signature of exhausted CD8+ T cells decreased over that time (Figure 6D). Gene set enrichment analysis of the PBMCs revealed reduced tumor necrosis factor-α (TNFA) signaling, especially in CD8+ T cells. In addition, differential expression analysis between T and NK cells before (C1, D1) and after treatment (C1, D8) revealed increased expression of cytotoxicity-associated genes (GZMK, GNLY) (Figure 6E). Cumulatively, this demonstrates peripheral antitumor immune system changes in response to treatment with WP1066. CD8+ T cells exhibited upregulation of gene sets associated with DNA repair, the G2/M cell cycle checkpoint, and apoptosis from before treatment to C1D8 of WP1066 treatment (Supplemental Figure 3). STAT3 activation is known to reduce immune cytotoxicity effector functions, and WP1066 treatment increased TNFA signaling in the PBMCs from before treatment to C1D8 of WP1066 treatment, suggesting increased cytotoxic immune cell activity. STAT3 activation is also known to suppress the activation of MPs and limit inflammatory responses in the TME. WP1066 treatment resulted in the upregulation of genes associated with regulation of inflammatory pathways (IFNG, HIF1A targets, NFKB) in myeloid cells from before treatment to C1D8 of WP1066 therapy (Figure 7A and Supplemental Figure 4). In addition, inflammation regulation (MAP3K8, NAMPT) (46, 47), stress (FKBP5) (48), and differentiation-related (MAFB) (49) genes were identified as significantly increasing in expression from before treatment to C1D8 (Figure 7B). Overall, this indicates a reduction in systemic immunosuppression within 8 days of treatment with WP1066, indicating that there are longer-term impacts on the immune response as a result of the transient decreases in p-STAT3 with each WP1066 dosing interval.
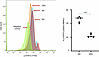 Figure 4
Figure 4WP1066 suppresses STAT3 activation in PBMCs. Left: Representative flow cytometric analysis of PBMCs from patient AflacST1901-04 during cycle 1, days 1 and 2, demonstrating decrease in phosphorylation (activation) of STAT3 [p-STAT3 (Y705)] at 4 hours and 24 hours after treatment with WP1066 (arrows labeled 4hr and 24hr) compared with baseline pretreatment levels (arrow labeled 0hr) and isotype IgG negative control for p-STAT3. Right: For all patients analyzed (AflacST1901-04, -05, -06, -07, -08), the plot shows a significant reduction in the median percentage of p-STAT3–positive PBMCs from cycle 1, day 1 (C1D1) at hour 0 prior to treatment (0hr) to cycle 1, day 2 (C1D2) at hour 24 after treatment (24hr) with WP1066. Wilcoxon’s rank sum tests were used to compare the percentage of p-STAT3–positive cells from C1D1 0-hour versus C1D2 24-hour time points. Median percentage positive cells is shown by horizontal solid bar; ***P < 0.002. Samples were run in experimental triplicates.
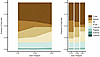 Figure 5
Figure 5WP1066 alters immune cell compartments in PBMCs. Shown are the proportion of total cells assigned to each cell type in the PBMC samples at each time point: stacked area (left) and bar plots (right). The y axis of both plots represents the number of cells labeled as a cell type at the specified time point, divided by the total number of cells for that time point. Shown is an increase in T and B cells from before WP1066 treatment to the start of cycle 2 of treatment with WP1066. PBMCs analyzed correspond to the PBMC samples analyzed by flow cytometry demonstrating suppressed p-STAT3 after WP1066 treatment (AflacST1901-04, -05, -06, -07, -08) and one additional subject (AflacST1901-010) who did not have flow cytometric analysis for PBMC p-STAT3. Only subject AflacST1901-010 received concomitant corticosteroids during treatment, and the dosage of steroids administered remained constant throughout. To identify the proportion of CD4+ and CD8+ T cells present in the PBMCs, module scores were calculated (Seurat v4) for all T cells and for different T cell subtype signatures from the literature (56). In addition, gene set enrichment analysis was performed to assess the underlying biological mechanisms active in the PBMCs at different time points. The escape package (60) was used to assess the enrichment of different hallmark gene sets (MSigDB).
 Figure 6
Figure 6Changes in T cell subsets in response to WP1066. CD4+ and CD8+ T cell subsets were estimated using phenotype signatures from the literature and assessing the expression of these signatures in T cell subsets in PBMCs. PBMCs analyzed correspond to the PBMC samples analyzed by flow cytometry demonstrating suppressed p-STAT3 after WP1066 treatment (AflacST1901-04, -05, -06, -07, -08) and one additional subject (AflacST1901-010) who did not have flow cytometric analysis for PBMC p-STAT3. Only subject AflacST1901-010 received concomitant corticosteroids during treatment, and the dosage of steroids administered remained constant throughout. Module score function from Seurat was used to represent the aggregate expression of these signatures in the cells. (A and B) Module scores for CD4+ (A) and CD8+ (B) naive signatures are shown for the designated time points. (C and D) Module scores for Treg signature (C) and CD8+ T cell exhaustion signature (D) are shown. Wilcoxon’s rank sum tests were used to compare module scores for cells from C1D1 versus C1D8 and C1D8 versus C2D1 time points. The resulting P values are shown: ****P < 0.0001; ns, not significant, P > 0.05. (E) Differential expression analysis was performed to identify genes significantly changing in expression in T and NK cells between C1D1 and C1D8 time points. Genes increasing after treatment (UP @ C1D8, purple) were identified based on the adjusted P value and average log2(fold change) [log2(FC)] results [adjusted P value < 0.05, average log2(FC) > 0.25]. DEG, differentially expressed gene.
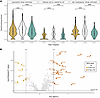 Figure 7
Figure 7The upregulated inflammatory pathways and markers in myeloid cells with WP1066 treatment. (A) Gene set enrichment analysis was performed to calculate the enrichment of the inflammatory pathway gene set in myeloid cells across time points. Gene sets were obtained from the MSigDB collection. Wilcoxon’s rank sum tests were used to compare enrichment scores among cells from C1D1 versus C1D8 and C1D8 versus C2D1 time points. Cell counts are shown in the figure legend. ****P < 0.0001. (B) Differential expression analysis was performed to identify genes significantly changing in expression in myeloid cells between C1D1 and C1D8 time points. Genes increasing after treatment (UP @ C1D8, orange) and decreasing after treatment (UP @ C1D1, gold) were identified based on the adjusted P value and average log2(fold change) [log2(FC)] results [adjusted P value < 0.05, |average log2(FC)| > 0.5]. DEG, differentially expressed gene.
-
Discussion
This phase I clinical trial was a first-in-child study of the orally administered BBB-penetrant p-STAT3 inhibitor WP1066. Here, we identified an MFD of 8 mg/kg on a Monday-Wednesday-Friday schedule for the first 14 days of each 28-day cycle in children with refractory brain tumors. Dose escalation beyond 8 mg/kg was stopped because patients expressed oral displeasure at this dose, and because 8 mg/kg was determined to be the phase II adult dose for WP1066 in combination with radiation therapy (RT) based on preclinical testing showing that allometric 8 mg/kg is optimal for promoting an antitumor immune response in mouse GBM models (22). It is worth noting that premedication with a viscous lidocaine mouth spray and antiemetics increased the oral tolerability of WP1066. Otherwise, the drug was well tolerated with minimal toxicity, confirming that administration of WP1066 in children with brain tumors is safe and feasible.
The median progression-free survival and overall survival, 1.8 and 4.9 months, respectively, are similar to the survival rates reported in the adult GBM phase I trial (45). Most notable was the one patient with DIPG who, after being taken off study for progression, continued to exhibit significant improvement in physical function for another 6 months off steroids and without further therapy, suggesting that the initial increase in tumor size may have been due to treatment-related pseudo-progression. Based on historical data, the timing and duration of this patient’s radiographic and clinical improvement with WP1066 treatment, which started more than 4 weeks after palliative re-RT, could be considered atypical for the clinical course following re-RT alone in a steroid-dependent patient with progressed DIPG (50). Likewise, among the H3.3G34R/V-mutant HGG patients treated with compassionate-use WP1066, one remained free of tumor progression for 17 months while on WP1066 treatment after completion of upfront RT. This clinical course is also atypical for H3.3G34R/V-mutant HGG, which historically has a median progression-free survival of about 10 months (51). No other patients reported here received RT within 4–6 weeks prior to starting WP1066. Together, these results suggest that WP1066 may have clinical benefit after RT in select brain tumor patients (i.e., STAT3-dependent tumors such as DIPG and H3.3G34R/V-mutant HGG) (9, 16, 17). Indeed, in preclinical models of GBM, it was not until RT was used in combination with WP1066 that a therapeutic response resulting in long-term survival and significantly enhanced median survival was observed (22). In that murine model, the efficacy of combining RT with WP1066 appeared to be due to RT-induced immune stimulation rather than RT-mediated cell killing, as this effect was lost in immune-incompetent mice.
PBMC p-STAT3 is upregulated in adult HGG patients (8) and thus could serve as a potential surrogate marker for STAT3 activity in the TME. In the adult GBM phase I trial, patients demonstrated suppression of PBMC p-STAT3 expression by flow cytometry, even at the lowest WP1066 dose tested (1 mg/kg), as early as 172 hours after treatment (45). In comparison, flow cytometric analysis of patient PBMCs in our trial demonstrated approximately 50% reduction in p-STAT3 expression 24 hours after the first dose of WP1066, and at all dose levels. Together these data confirm that WP1066 is effective in suppressing systemic STAT3 activity. Although subsequent dosing 7 days after the first dose given showed similar decreases in p-STAT3, we did not observe a prolonged or cumulative effect, as PBMC p-STAT3 levels had reverted to baseline before the second week of drug administration. This was the anticipated result given that PBMCs are being continuously replenished between the Monday-Wednesday-Friday weekly dosing intervals. One limitation of our study is that we were not able to correlate these changes in PBMCs with TME p-STAT3 expression without immediate pre- and post-treatment tumor tissue to analyze. Analysis of PBMCs and tumor tissue before and after treatment with WP1066 is planned for the current adult phase II clinical trial of WP1066 in GBM patients (NCT05879250) and for future pediatric trials.
A second limitation of our study is that we were not able to obtain reliable pharmacokinetic (PK) data for WP1066. The adult phase I trial in GBM patients initially reported PK data for WP1066 using a preliminary assay that failed to accurately estimate available WP1066 (45). This is in part a result of the unique properties of WP1066, a Michael acceptor that reacts reversibly with nucleophiles, especially sulfhydryl groups of amino acids. WP1066 was formulated to allow for extended release, but it does not follow pharmacokinetics of the naked drug. Despite investigation by multiple laboratories, no method currently exists for measuring free and active forms of WP1066. Although we collected samples for PK analysis in our study, and analyzed the first 3 patients, the results were highly variable, and thus we did not analyze the remaining samples. Further confounding the assay’s accuracy is that PK data are inherently more variable with oral versus intravenous drug dosing. Optimization of the assay is currently ongoing.
Corresponding to the reduction in PBMC p-STAT3 with WP1066 treatment, we observed specific alterations in the proportion and function of peripheral circulating immune cells toward an antitumor immune response, confirming the anti-STAT3 functional activity of WP1066 treatment in this patient population. Indeed, consistent with the preclinical mechanism of action described for WP1066 (52–54), we observed PBMC gene signatures after treatment reflective of increased CD4+ and CD8+ T cells, increased proinflammatory TNFA signaling, increased inflammatory, stress, and differentiation activity in myeloid cells, and downregulation of Tregs. Notably, while the decreased PBMCs’ p-STAT3 levels reverted to baseline between dosing intervals, there appear to be longer-term downstream effects as evidenced by the immune cell changes observed by RNA-seq analysis at cycle 2, hour 0. However, there did not appear to be a dose-dependent difference between the RNA-seq analyses performed at dose level 2 versus dose level 3. Importantly, these changes were unrelated to concomitant steroid use, as only one patient analyzed was receiving steroids during therapy and the steroid dosage was stable throughout treatment. Whether these peripheral changes reflect shifts in immune cells trafficking to the TME is again unclear without pre- and post-treatment tissue to analyze. However, studies have shown that ablating STAT3 only in murine hematopoietic cells results in marked enhancement of T, NK, and dendritic cell function in tumor-bearing mice and potent local antitumor effects in vivo (28). Thus, it is conceivable that the systemic immune responses induced by WP1066 treatment may be sufficient to generate an antitumor response within the TME, irrespective of the amount of WP1066 that crosses the BBB. Although we observed systemic antitumor immune responses over a short treatment interval, all patients progressed in a relatively short time on therapy. Historically, single-agent immunotherapy, such as immune checkpoint inhibitors, typically requires weeks to months to generate an effective antitumor response (55), and thus it may be unrealistic to expect a significant impact on survival in the context of a phase I study. However, based on our trial results showing both drug activity and possible clinical benefit, further investigation of the efficacy of WP1066 treatment in these patient populations, especially with RT and possibly in the upfront setting for pHGG, is warranted.
-
Methods
Sex as a biological variable. Our study examined male and female pediatric patients, and no differences in safety profile were detected between the sexes, but only 1 female was enrolled. Thus, the study was not powered to confirm safety profile differences based on sex.
Study design and participants. We performed an open-label, single-center, single-arm, 3+3 dose escalation phase I clinical trial design in pediatric patients with primary malignant brain tumors that had progressed or recurred after standard therapy (NCT04334863). The study was conducted at the Aflac Cancer and Blood Disorders Center of Children’s Healthcare of Atlanta under an IRB-approved protocol (AflacST1901). Patients were between the ages of 3 and 25 years, had a Karnofsky or Lansky Performance Scale score of 60 or higher, had previously undergone standard-of-care treatment including surgery, radiation, and/or first-line adjuvant chemotherapy, and had magnetic resonance imaging (MRI) evidence of recurrent or progressive tumor. No single lesion could be larger than 5 cm in maximal diameter. There was no restriction on the number of prior episodes of tumor progression or treatments given. To be eligible for enrollment, a minimum of a 3-week washout was required from the patient’s last dose of myelosuppressive chemotherapy, or 6 weeks from the last dose of nitrosourea-containing chemotherapy. Patients must have received their last dose of an anticancer investigational or biologic drug more than 7 days before enrollment, 21 days before their last antibody therapy, or 42 days before their last immunotherapy. Patients were also required to be more than 12 weeks from autologous stem cell transplantation. Craniospinal radiation must have been completed at least 3 months before enrollment. Other bone marrow irradiation must have been completed at least 6 weeks before and local palliative irradiation at least 2 weeks before enrollment. Patients being treated with high-dose corticosteroids were required to have received a stable or decreasing dose for at least 1 week before enrollment.
A separate cohort of 3 young adult patients with supratentorial H3.3G34R/V-mutant HGG were treated off-study with compassionate-use WP1066 (provided by Moleculin Inc., Houston, Texas, USA) at the University of Michigan C.S. Mott Children’s Hospital, after having received prior treatment with at least standard-of-care radiation therapy and temozolomide chemotherapy. One of these 3 patients was treated with WP1066 and concurrent adjuvant temozolomide after completing standard radiation and had no evidence of disease at the time of WP1066 initiation. While the compassionate-use patients do not contribute to the pediatric phase I data since these data relate to identifying the maximum tolerated dose and describing the safety and tolerability of WP1066 in this population, they are included here because they enrich for the HGG population — in particular, H3G34R tumors that have been shown preclinically to be dependent on STAT3 activity. Thus, data from these patients may provide further information about the preliminary efficacy of WP1066.
Study treatment. Eligible patients were assigned to a dose of WP1066 using a 3+3 design algorithm. On-study patients were treated according to the adult HGG regimen with WP1066 administered orally twice daily on Monday, Wednesday, and Friday for the first 14 days in each 28-day treatment cycle. Patients received WP1066 as a powder based on their weight. Immediately before administration, WP1066 was reconstituted into a pharmacy-prepared peppermint-bubblegum-flavored liquid suspension (mixing of the study drug with additional flavored syrups or other liquid additives to improve taste and palatability for children was not allowed because of unknown biocompatibility). Patients could receive WP1066 for up to 1 year (12 cycles) or until disease progression or unacceptable toxicity. The starting dose of WP1066 was 4 mg/kg, which was the tolerated dose in the adult trial of WP1066 at the time of initiation of our pediatric trial with WP1066 (NCT04334863). The planned dosing regimens for investigation were 4, 6, 8, and 16 mg/kg.
Before treatment was advanced to the next dose level, a cohort summary was discussed with the clinical research monitor and approved by a data safety monitoring board. After a baseline MRI, treatment responses were assessed by MRIs every 2–3 months, or earlier if clinically indicated to rule out progression. Laboratory parameters were measured at baseline and before the start of each treatment cycle. Correlative biology blood samples for flow cytometric analysis of PBMC p-STAT3 and PBMC single-cell RNA sequencing for immunophenotyping were obtained before WP1066 treatment and at scheduled time points after the start of treatment. Adverse events were graded according to the National Cancer Institute’s Common Terminology Criteria for Adverse Events, version 5.0.
Flow cytometry for PBMC p-STAT3 expression. Flow cytometry for activated p-STAT3 was performed as previously described (45). PBMCs were obtained from patients on cycle 1 day 1 (C1D1) at 0 hours and 4 hours, C1D2 at 0 hours, and C1D8 at 0 hours and 4 hours. Because the p-STAT3 signal is rapidly lost ex vivo, the blood was immediately processed and analyzed. PBMCs were isolated from heparin blood by density gradient centrifugation with Histopaque Ficoll 1077 (Sigma-Aldrich) and counted. Six million cells were then resuspended in 0.6 mL of PBS. Paraformaldehyde (Thermo Fisher Scientific) was pre-warmed at 37°C and added to achieve a final concentration of 2%. The solution was then incubated for 10 minutes at 37°C, chilled on ice for 1 minute, and equally distributed into 5 wells of a 96-well U-bottom plate. For permeabilization, the paraformaldehyde was removed by pelleting of the cells at 700g for 5 minutes, resuspension of the cells in pre-chilled 90% methanol, and incubation on ice for 30 minutes. The cells were then pelleted at 700g for 5 minutes at 4°C, washed with FACS staining buffer (Thermo Fisher Scientific), and again pelleted at 700g for 5 minutes at 4°C. Two wells were then resuspended in staining buffer including the PE-labeled p-STAT3 (Y705) antibody, two wells in staining buffer including isotype control antibody, and one well in staining buffer only. After 1 hour of incubation at room temperature, the cells were washed again with FACS staining buffer before being transferred to FACS tubes for the flow analysis. Duplicate specimens were processed in parallel.
scRNA-seq. scRNA-seq was performed on PBMC samples retrieved at C1D1, C1D8, and C2D1 time points. Gene expression libraries were generated and barcoded using the 10x Genomics Chromium Single-Cell 5′ kit and sequenced using the NovaSeq 6000 platform. Cell Ranger v7 was used to align FASTQs to the transcriptome (GRCh38-2020-A), with mode introns=False and Single Cell 5′ PE chemistry options specified. Aligned cell count matrices for each of the 17 samples were merged and analyzed in downstream steps using Seurat v4 functions (56). The dataset was filtered to identify high-quality cells by keeping cells with low mitochondrial content (<10%) and feature counts in the normal range (>300, <2,500). In addition, we filtered out technical doublets using DoubletFinder v2 (57), resulting in 62,000 high-quality cells for further analysis. SCTransform from Seurat was used for the normalization and scaling of counts, and then principal component analysis, neighborhood analysis, clustering, and uniform manifold approximation and projection (UMAP) analysis were performed to identify unsupervised clusters and project the cells to a 2-dimensional embedding for easier interpretation. Upon examining the UMAPs and clustering, we determined there was no batch effect (Supplemental Figure 5), so we did not use the integration and harmonization methods to overcorrect data. SingleR (58) and canonical cell type markers were used to annotate clusters.
Statistics. Patient demographic and clinical characteristics were summarized using descriptive statistics. Dose-limiting toxicities were counted according to dose levels, while toxicities within the evaluable patient cohort were categorized and summarized by grade. Progression-free survival and overall survival were analyzed using Kaplan-Meier method. The statistical analyses were performed using R software, version 4.3.0. To identify the proportion of CD4+ and CD8+ T cells present in the PBMCs, we calculated the module scores (Seurat v4) for all T cells and for different T cell subtype signatures from the literature (59). In addition, gene set enrichment analysis was performed to assess the underlying biological mechanisms active in the PBMCs at different time points. The escape package (60) was used to assess the enrichment of different hallmark gene sets (Molecular Signatures Database [MSigDB]). Wilcoxon’s rank sum tests were used to compare gene expression, module scores, enrichment scores, and cell type proportions between cells or samples from C1D1 versus C1D8 and C1D8 versus C2D1 time points. While we were limited by sample size per time point, we performed a pseudobulk approach when validating the changes in signatures represented in Figures 5 and 6. First, we summed counts for each gene across all the cells in each unique combination of patient and time point (combining replicates). Then, we performed an RNA-seq analysis approach using DESeq2 methodology, including filtering of genes with low counts, estimation of size factors, and normalization. Then, we performed gene set variation analysis (using the GSVA package) treating each patient as an observation within each time point, comparing enrichment scores across time points.
Study approval. This study was approved by the Institutional Review Boards of Emory University and Children’s Healthcare of Atlanta before study initiation, and written informed signed consent was obtained from study participants aged 18 years or older or from the legal guardian(s) of study participants less than 18 years of age prior to any study procedures or participation.
Data availability. After lockout of the database, all generated data are now available through the Pediatric Cancer Data Commons portal (https://commons.cri.uchicago.edu/pcdc/) and are available in the Supporting Data Values file.
-
Author contributions
TJM, ABH, WP, RCC, DA, and RK were responsible for the primary study design. TJM, RCC, and DA conducted the study. HM and MB performed the sequencing and analysis. BY, HR, and KD assisted in flow cytometry analysis. MS provided neuropathology support. ZH performed the clinical data analysis. RCC, TJM, HM, AF, and ZH drafted the manuscript, and ABH, WP, and DA read the draft manuscript for detailed editorial feedback. All authors reviewed and approved the manuscript.
-
Funding support
The following entities provided funding support.
- CURE Childhood Cancer (to TJM).
- Peach Bowl Inc. (to TJM).
-
Acknowledgments
The team acknowledges the stellar work of the Clinical Research Office of the Aflac Cancer and Blood Disorders Center of Children’s Healthcare of Atlanta, and the entire team at Moleculin Inc. for providing the WP1066 investigational agent and overall support for the study. ABH currently holds NIH grants related to the investigation of WP1066.
Address correspondence to: Tobey J. MacDonald, 1760 Haygood Drive NE, Health Sciences Research Building, Room E384, Atlanta, Georgia 30322, USA. Phone: 404.727.1447; Email: tobey.macdonald@emory.edu.
-
Footnotes
Conflict of interest: WP is the inventor of WP1066 and is on the advisory board of and has a grant from Moleculin Inc.
Copyright: © 2025, Castellino et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2026;11(3):e194823.https://doi.org/10.1172/jci.insight.194823.
-
References
- MacDonald TJ, et al. Treatment of high-grade glioma in children and adolescents. Neuro Oncol. 2011;13(10):1049–1058.
- Baugh JN, et al. Treatment-related survival patterns in diffuse intrinsic pontine glioma using a historical cohort: a report from the European Society for Pediatric Oncology DIPG/DMG registry. Neurooncol Adv. 2024;6(1):vdae155. View this article via: PubMed Google Scholar
- Zaghloul MS, et al. Re-irradiation for recurrent/progressive pediatric brain tumors: from radiobiology to clinical outcomes. Expert Rev Anticancer Ther. 2023;23(7):709–717.
- Slika H, et al. Overcoming treatment resistance in medulloblastoma: underlying mechanisms and potential strategies. Cancers (Basel). 2024;16(12):2249.
- Phi JH, et al. Overcoming chemoresistance of pediatric ependymoma by inhibition of STAT3 signaling. Transl Oncol. 2015;8(5):376–386.
- Kumar S, et al. Inhibition of STAT3: a promising approach to enhancing the efficacy of chemotherapy in medulloblastoma. Transl Oncol. 2024;46:102023.
- Rahaman SO, et al. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21(55):8404–8413.
- Humphries W, et al. Detecting the percent of peripheral blood mononuclear cells displaying p-STAT-3 in malignant glioma patients. J Transl Med. 2009;7:92.
- Park J, et al. STAT3 is a key molecule in the oncogenic behavior of diffuse intrinsic pontine glioma. Oncol Lett. 2020;20(2):1989–1998.
- Doucette TA, et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012;14(9):1136–1145.
- Carpenter RL, Lo HW. STAT3 target genes relevant to human cancers. Cancers (Basel). 2014;6(2):897–925.
- Fu W, et al. Roles of STAT3 in the pathogenesis and treatment of glioblastoma. Front Cell Dev Biol. 2023;11:1098482.
- Abou-Ghazal M, et al. The incidence, correlation with tumor-infiltrating inflammation, and prognosis of phosphorylated STAT3 expression in human gliomas. Clin Cancer Res. 2008;14(24):8228–8235.
- Tan MSY, et al. A STAT3-based gene signature stratifies glioma patients for targeted therapy. Nat Commun. 2019;10(1):3601.
- Yu H, et al. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7(1):41–51.
- Anderson JB, et al. STAT3 as a biologically relevant target in H3K27M-mutant diffuse midline glioma. Oncotarget. 2023;14:858–859.
- Sweha SR, et al. Epigenetically defined therapeutic targeting in H3.3G34R/V high-grade gliomas. Sci Transl Med. 2021;13(615):eabf7860.
- Sherry-Lynes MM, et al. Regulation of the JMJD3 (KDM6B) histone demethylase in glioblastoma stem cells by STAT3. PLoS One. 2017;12(4):e0174775.
- Kesanakurti D, et al. Essential role of cooperative NF-κB and Stat3 recruitment to ICAM-1 intronic consensus elements in the regulation of radiation-induced invasion and migration in glioma. Oncogene. 2013;32(43):5144–5155.
- Halliday J, et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A. 2014;111(14):5248–5253.
- Lau J, et al. STAT3 blockade inhibits radiation-induced malignant progression in glioma. Cancer Res. 2015;75(20):4302–4311.
- Ott M, et al. Radiation with STAT3 blockade triggers dendritic cell-T cell interactions in the glioma microenvironment and therapeutic efficacy. Clin Cancer Res. 2020;26(18):4983–4994.
- Zaiter A, et al. STAT3 in medulloblastoma: a key transcriptional regulator and potential therapeutic target. Mol Biol Rep. 2022;49(11):10635–10652.
- Yuan L, et al. STAT3 is required for Smo-dependent signaling and mediates Smo-targeted treatment resistance and tumorigenesis in Shh medulloblastoma. Mol Oncol. 2022;16(4):1009–1025.
- Wu A, et al. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12(11):1113–1125.
- Wang T, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10(1):48–54.
- O’Farrell AM, et al. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J. 1998;17(4):1006–1018.
- Kortylewski M, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11(12):1314–1321.
- Zorn E, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579.
- Lin EY, et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006;66(23):11238–11246.
- Hussain SF, et al. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007;67(20):9630–9636.
- Verstovsek S, et al. WP1066, a novel JAK2 inhibitor, suppresses proliferation and induces apoptosis in erythroid human cells carrying the JAK2 V617F mutation. Clin Cancer Res. 2008;14(3):788–796.
- Iwamaru A, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26(17):2435–2444.
- Horiguchi A, et al. STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell carcinoma. Br J Cancer. 2010;102(11):1592–1599.
- Lee HT, et al. Stat3 orchestrates interaction between endothelial and tumor cells and inhibition of Stat3 suppresses brain metastasis of breast cancer cells. Oncotarget. 2015;6(12):10016–10029.
- Kupferman ME, et al. Therapeutic suppression of constitutive and inducible JAK\STAT activation in head and neck squamous cell carcinoma. J Exp Ther Oncol. 2009;8(2):117–127.View this article via: PubMed Google Scholar
- Lin L, et al. A novel small molecule, LLL12, inhibits STAT3 phosphorylation and activities and exhibits potent growth-suppressive activity in human cancer cells. Neoplasia. 2010;12(1):39–50.
- Tsujita Y, et al. STAT3 inhibition by WP1066 suppresses the growth and invasiveness of bladder cancer cells. Oncol Rep. 2017;38(4):2197–2204.
- Geng L, et al. WP1066 exhibits antitumor efficacy in nasal-type natural killer/T-cell lymphoma cells through downregulation of the STAT3 signaling pathway. Oncol Rep. 2016;36(5):2868–2874.
- Judd LM, et al. Inhibition of the JAK2/STAT3 pathway reduces gastric cancer growth in vitro and in vivo. PLoS One. 2014;9(5):e95993.
- Kong LY, et al. A novel inhibitor of signal transducers and activators of transcription 3 activation is efficacious against established central nervous system melanoma and inhibits regulatory T cells. Clin Cancer Res. 2008;14(18):5759–5768.
- Kong LY, et al. Intratumoral mediated immunosuppression is prognostic in genetically engineered murine models of glioma and correlates to immunotherapeutic responses. Clin Cancer Res. 2010;16(23):5722–5733.
- Assi HH, et al. Preclinical characterization of signal transducer and activator of transcription 3 small molecule inhibitors for primary and metastatic brain cancer therapy. J Pharmacol Exp Ther. 2014;349(3):458–469.
- Heimberger AB, et al. The role of tregs in human glioma patients and their inhibition with a novel STAT-3 inhibitor. Clin Neurosurg. 2009;56:98–106.View this article via: PubMed Google Scholar
- Groot J, et al. A first-in-human phase I trial of the oral p-STAT3 inhibitor WP1066 in patients with recurrent malignant glioma. CNS Oncol. 2022;11(2):CNS87.
- Xu D, et al. TPL2 kinase action and control of inflammation. Pharmacol Res. 2018;129:188–193.
- Audrito V, et al. NAMPT and NAPRT: two metabolic enzymes with key roles in inflammation. Front Oncol. 2020;10:358.
- Zannas AS, et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB-driven inflammation and cardiovascular risk. Proc Natl Acad Sci U S A. 2019;116(23):11370–11379.
- Kelly LM, et al. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19(9):1987–1997.
- Lu VM, et al. Reirradiation for diffuse intrinsic pontine glioma: a systematic review and meta-analysis. Childs Nerv Syst. 2019;35(5):739–746.
- Crowell C, et al. Systematic review of diffuse hemispheric glioma, H3 G34-mutant: outcomes and associated clinical factors. Neurooncol Adv. 2022;4(1):vdac133. View this article via: PubMed Google Scholar
- Wei J, et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther. 2010;9(1):67–78.
- Hatiboglu MA, et al. Immune therapeutic targeting of glioma cancer stem cells. Target Oncol. 2010;5(3):217–227.
- Masliantsev K, et al. Impact of STAT3 phosphorylation in glioblastoma stem cells radiosensitization and patient outcome. Oncotarget. 2017;9(3):3968–3979.
- Yin J, et al. What is the optimal duration of immune checkpoint inhibitors in malignant tumors? Front Immunol. 2022;13:983581.
- Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.
- McGinnis CS, et al. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337.
- Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163–172.
- Chu Y, et al. Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat Med. 2023;29(6):1550–1562.
- Borcherding N, Andrews J. Easy single cell analysis platform for enrichment. Version 1.6.0. https://s3.jcloud.sjtu.edu.cn/899a892efef34b1b944a19981040f55b-oss01/bioconductor/3.15/bioc/manuals/escape/man/escape.pdf Accessed December 17, 2025.
-
Version history
- Version 1 (December 11, 2025): In-Press Preview
- Version 2 (February 9, 2026): Electronic publication













